Co-expression Networks From Gene Expression Variability Between Genetically Identical Seedlings Can Reveal Novel Regulatory Relationships
- PMID: 33384705
- PMCID: PMC7770228
- DOI: 10.3389/fpls.2020.599464
Co-expression Networks From Gene Expression Variability Between Genetically Identical Seedlings Can Reveal Novel Regulatory Relationships
Abstract
Co-expression networks are a powerful tool to understand gene regulation. They have been used to identify new regulation and function of genes involved in plant development and their response to the environment. Up to now, co-expression networks have been inferred using transcriptomes generated on plants experiencing genetic or environmental perturbation, or from expression time series. We propose a new approach by showing that co-expression networks can be constructed in the absence of genetic and environmental perturbation, for plants at the same developmental stage. For this, we used transcriptomes that were generated from genetically identical individual plants that were grown under the same conditions and for the same amount of time. Twelve time points were used to cover the 24-h light/dark cycle. We used variability in gene expression between individual plants of the same time point to infer a co-expression network. We show that this network is biologically relevant and use it to suggest new gene functions and to identify new targets for the transcriptional regulators GI, PIF4, and PRR5. Moreover, we find different co-regulation in this network based on changes in expression between individual plants, compared to the usual approach requiring environmental perturbation. Our work shows that gene co-expression networks can be identified using variability in gene expression between individual plants, without the need for genetic or environmental perturbations. It will allow further exploration of gene regulation in contexts with subtle differences between plants, which could be closer to what individual plants in a population might face in the wild.
Keywords: Arabidopsis; co-expression analysis; gene expression; modules; networks; seedlings; variability.
Copyright © 2020 Cortijo, Bhattarai, Locke and Ahnert.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


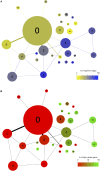

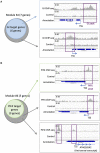
References
-
- Blondel V. D., Guillaume J.-L., Lambiotte R., Lefebvre E. (2008). Fast unfolding of communities in large networks. J. Stat. Mech. 2008:10008 10.1088/1742-5468/2008/10/P10008 - DOI
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

