A Pilot Study of the Effect of Deployment on the Gut Microbiome and Traveler's Diarrhea Susceptibility
- PMID: 33384968
- PMCID: PMC7770225
- DOI: 10.3389/fcimb.2020.589297
A Pilot Study of the Effect of Deployment on the Gut Microbiome and Traveler's Diarrhea Susceptibility
Abstract
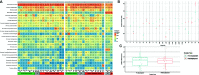
Traveler's diarrhea (TD) is a recurrent and significant issue for many travelers including the military. While many known enteric pathogens exist that are causative agents of diarrhea, our gut microbiome may also play a role in TD susceptibility. To this end, we conducted a pilot study of the microbiome of warfighters prior to- and after deployment overseas to identify marker taxa relevant to TD. This initial study utilized full-length 16S rRNA gene sequencing to provide additional taxonomic resolution toward identifying predictive taxa.16S rRNA analyses of pre- and post-deployment fecal samples identified multiple marker taxa as significantly differentially abundant in subjects that reported diarrhea, including Weissella, Butyrivibrio, Corynebacterium, uncultivated Erysipelotrichaceae, Jeotgallibaca, unclassified Ktedonobacteriaceae, Leptolinea, and uncultivated Ruminiococcaceae. The ability to identify TD risk prior to travel will inform prevention and mitigation strategies to influence diarrhea susceptibility while traveling.
Keywords: 16S rRNA gene; deployment; diarrhea; dysbiosis; gut microbiome.
Copyright © 2020 Stamps, Lyon, Irvin, Kelley-Loughnane and Goodson.
Conflict of interest statement
BS is an employee of UES, Inc. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Abriouel H., Lerma L. L., Casado Muñoz M., del C., Montoro B. P., Kabisch J., et al. (2015). The controversial nature of the Weissella genus: technological and functional aspects versus whole genome analysis-based pathogenic potential for their application in food and health. Front. Microbiol. 6, 1–14. 10.3389/fmicb.2015.01197 - DOI - PMC - PubMed
-
- Andersen K. S., Kirkegaard R. H., Karst S. M., Albertsen M. (2018). ampvis2: an R package to analyse and visualise 16S rRNA amplicon data. bioRxiv 299537, 1–2. 10.1101/299537 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

