An Eight Amino Acid Segment Controls Oligomerization and Preferred Conformation of the two Non-visual Arrestins
- PMID: 33387531
- PMCID: PMC7870585
- DOI: 10.1016/j.jmb.2020.166790
An Eight Amino Acid Segment Controls Oligomerization and Preferred Conformation of the two Non-visual Arrestins
Abstract
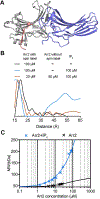
G protein coupled receptors signal through G proteins or arrestins. A long-standing mystery in the field is why vertebrates have two non-visual arrestins, arrestin-2 and arrestin-3. These isoforms are ~75% identical and 85% similar; each binds numerous receptors, and appear to have many redundant functions, as demonstrated by studies of knockout mice. We previously showed that arrestin-3 can be activated by inositol-hexakisphosphate (IP6). IP6 interacts with the receptor-binding surface of arrestin-3, induces arrestin-3 oligomerization, and this oligomer stabilizes the active conformation of arrestin-3. Here, we compared the impact of IP6 on oligomerization and conformational equilibrium of the highly homologous arrestin-2 and arrestin-3 and found that these two isoforms are regulated differently. In the presence of IP6, arrestin-2 forms "infinite" chains, where each promoter remains in the basal conformation. In contrast, full length and truncated arrestin-3 form trimers and higher-order oligomers in the presence of IP6; we showed previously that trimeric state induces arrestin-3 activation (Chen et al., 2017). Thus, in response to IP6, the two non-visual arrestins oligomerize in different ways in distinct conformations. We identified an insertion of eight residues that is conserved across arrestin-2 homologs, but absent in arrestin-3 that likely accounts for the differences in the IP6 effect. Because IP6 is ubiquitously present in cells, this suggests physiological consequences, including differences in arrestin-2/3 trafficking and JNK3 activation. The functional differences between two non-visual arrestins are in part determined by distinct modes of their oligomerization. The mode of oligomerization might regulate the function of other signaling proteins.
Keywords: IP(6); isoforms; oligomer; signaling protein; structure.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures





References
-
- Wilden U (1995). Duration and amplitude of the light-induced cGMP hydrolysis in vertebrate photoreceptors are regulated by multiple phosphorylation of rhodopsin and by arrestin binding. Biochemistry 34, 1446–54. - PubMed
-
- Krupnick JG, Gurevich VV & Benovic JL (1997). Mechanism of quenching of phototransduction. Binding competition between arrestin and transducin for phosphorhodopsin. J Biol Chem 272, 18125–31. - PubMed
-
- Goodman OB Jr., Krupnick JG, Santini F, Gurevich VV, Penn RB, Gagnon AW, Keen JH & Benovic JL (1996). Beta-arrestin acts as a clathrin adaptor in endocytosis of the beta2-adrenergic receptor. Nature 383, 447–50. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

