A Fifth of the Protein World: Rossmann-like Proteins as an Evolutionarily Successful Structural unit
- PMID: 33387532
- PMCID: PMC7870570
- DOI: 10.1016/j.jmb.2020.166788
A Fifth of the Protein World: Rossmann-like Proteins as an Evolutionarily Successful Structural unit
Abstract
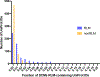
The Rossmann-like fold is the most prevalent and diversified doubly-wound superfold of ancient evolutionary origin. Rossmann-like domains are present in a variety of metabolic enzymes and are capable of binding diverse ligands. Discerning evolutionary relationships among these domains is challenging because of their diverse functions and ancient origin. We defined a minimal Rossmann-like structural motif (RLM), identified RLM-containing domains among known 3D structures (20%) and classified them according to their homologous relationships. New classifications were incorporated into our Evolutionary Classification of protein Domains (ECOD) database. We defined 156 homology groups (H-groups), which were further clustered into 123 possible homology groups (X-groups). Our analysis revealed that RLM-containing proteins constitute approximately 15% of the human proteome. We found that disease-causing mutations are more frequent within RLM domains than within non-RLM domains of these proteins, highlighting the importance of RLM-containing proteins for human health.
Keywords: Rossmann-fold; domains classification; minimal Rossmann-like motif; protein evolution.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


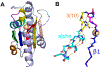
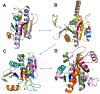
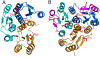


Similar articles
-
Functional analysis of Rossmann-like domains reveals convergent evolution of topology and reaction pathways.PLoS Comput Biol. 2019 Dec 23;15(12):e1007569. doi: 10.1371/journal.pcbi.1007569. eCollection 2019 Dec. PLoS Comput Biol. 2019. PMID: 31869345 Free PMC article.
-
Functional and evolutionary analysis of viral proteins containing a Rossmann-like fold.Protein Sci. 2018 Aug;27(8):1450-1463. doi: 10.1002/pro.3438. Epub 2018 Jun 13. Protein Sci. 2018. PMID: 29722076 Free PMC article.
-
Bridging the Gap between Sequence and Structure Classifications of Proteins with AlphaFold Models.J Mol Biol. 2024 Nov 15;436(22):168764. doi: 10.1016/j.jmb.2024.168764. Epub 2024 Aug 26. J Mol Biol. 2024. PMID: 39197652
-
Classification of proteins with shared motifs and internal repeats in the ECOD database.Protein Sci. 2016 Jul;25(7):1188-203. doi: 10.1002/pro.2893. Epub 2016 Feb 21. Protein Sci. 2016. PMID: 26833690 Free PMC article. Review.
-
Using the folding landscapes of proteins to understand protein function.Curr Opin Struct Biol. 2016 Feb;36:67-74. doi: 10.1016/j.sbi.2016.01.001. Epub 2016 Jan 24. Curr Opin Struct Biol. 2016. PMID: 26812092 Review.
Cited by
-
Structural characterization of two prototypical repressors of SorC family reveals tetrameric assemblies on DNA and mechanism of function.Nucleic Acids Res. 2024 Jul 8;52(12):7305-7320. doi: 10.1093/nar/gkae434. Nucleic Acids Res. 2024. PMID: 38842936 Free PMC article.
-
CryoEM structure of Rv2531c reveals cofactor-induced tetramer-dimer transition in a tuberculin amino acid decarboxylase.J Biol Chem. 2025 Jun 19;301(8):110394. doi: 10.1016/j.jbc.2025.110394. Online ahead of print. J Biol Chem. 2025. PMID: 40543586 Free PMC article.
-
Biological Catalysis and Information Storage Have Relied on N-Glycosyl Derivatives of β-D-Ribofuranose since the Origins of Life.Biomolecules. 2023 Apr 30;13(5):782. doi: 10.3390/biom13050782. Biomolecules. 2023. PMID: 37238652 Free PMC article. Review.
-
Creative destruction: New protein folds from old.Proc Natl Acad Sci U S A. 2022 Dec 27;119(52):e2207897119. doi: 10.1073/pnas.2207897119. Epub 2022 Dec 19. Proc Natl Acad Sci U S A. 2022. PMID: 36534803 Free PMC article.
-
Architecture, Function, Regulation, and Evolution of α-Glucans Metabolic Enzymes in Prokaryotes.Chem Rev. 2024 Apr 24;124(8):4863-4934. doi: 10.1021/acs.chemrev.3c00811. Epub 2024 Apr 12. Chem Rev. 2024. PMID: 38606812 Free PMC article. Review.
References
-
- Aravind L, Anantharaman V, & Koonin EV (2002). Monophyly of class I aminoacyl tRNA synthetase, USPA, ETFP, photolyase, and PP‐ATPase nucleotide‐binding domains: implications for protein evolution in the RNA world. Proteins. 48(1), 1–14. - PubMed
-
- Orengo CA, Jones DT, & Thornton JM (1994). Protein superfamilles and domain superfolds. Nature. 372(6507), 631–634. - PubMed
-
- Rossmann MG, Moras D, & Olsen KW (1974). Chemical and biological evolution of a nucleotide-binding protein. Nature. 250(5463), 194–199. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

