The Effect of SMN Gene Dosage on ALS Risk and Disease Severity
- PMID: 33389754
- PMCID: PMC8048961
- DOI: 10.1002/ana.26009
The Effect of SMN Gene Dosage on ALS Risk and Disease Severity
Abstract
Objective: The role of the survival of motor neuron (SMN) gene in amyotrophic lateral sclerosis (ALS) is unclear, with several conflicting reports. A decisive result on this topic is needed, given that treatment options are available now for SMN deficiency.
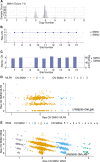
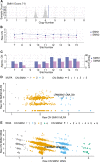
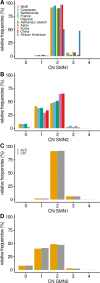
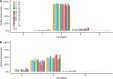
Methods: In this largest multicenter case control study to evaluate the effect of SMN1 and SMN2 copy numbers in ALS, we used whole genome sequencing data from Project MinE data freeze 2. SMN copy numbers of 6,375 patients with ALS and 2,412 controls were called from whole genome sequencing data, and the reliability of the calls was tested with multiplex ligation-dependent probe amplification data.
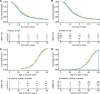
Results: The copy number distribution of SMN1 and SMN2 between cases and controls did not show any statistical differences (binomial multivariate logistic regression SMN1 p = 0.54 and SMN2 p = 0.49). In addition, the copy number of SMN did not associate with patient survival (Royston-Parmar; SMN1 p = 0.78 and SMN2 p = 0.23) or age at onset (Royston-Parmar; SMN1 p = 0.75 and SMN2 p = 0.63).
Interpretation: In our well-powered study, there was no association of SMN1 or SMN2 copy numbers with the risk of ALS or ALS disease severity. This suggests that changing SMN protein levels in the physiological range may not modify ALS disease course. This is an important finding in the light of emerging therapies targeted at SMN deficiencies. ANN NEUROL 2021;89:686-697.
© 2021 The Authors. Annals of Neurology published by Wiley Periodicals LLC on behalf of American Neurological Association.
Conflict of interest statement
X.C. and M.A.E. are employees of Illumina Inc.
Figures
References
-
- Hagenacker T, Wurster CD, Günther R, et al. Nusinersen in adults with 5q spinal muscular atrophy: a non‐interventional, multicentre, observational cohort study. Lancet Neurol 2020;19:317–325. - PubMed
-
- Pupillo E, Messina P, Logroscino G, et al. Long‐term survival in amyotrophic lateral sclerosis: a population‐based study. Ann Neurol 2014;75:287–297. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

