SARS-CoV-2 variants evolved during the early stage of the pandemic and effects of mutations on adaptation in Wuhan populations
- PMID: 33390836
- PMCID: PMC7757051
- DOI: 10.7150/ijbs.47827
SARS-CoV-2 variants evolved during the early stage of the pandemic and effects of mutations on adaptation in Wuhan populations
Abstract
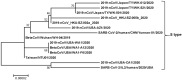
The outbreak of the coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The pandemic apparently started in December 2019 in Wuhan, China, and has since affected many countries worldwide, turning into a major global threat. Chinese researchers reported that SARS-CoV-2 could be classified into two major variants. They suggest that investigating the variations and characteristics of these variants might help assess risks and develop better treatment and prevention strategies. The two variants were named L-type and S-type, in which L-type was prevailed in an initial outbreak in Wuhan, Central China's Hubei Province, and S-type was phylogenetically older than L-type and less prevalent at an early stage, but with a later increase in frequency in Wuhan. There were 149 mutations in 103 sequenced SARS-CoV-2 genomes, 83 of which were nonsynonymous, leading to alteration in the amino acid sequence of proteins. Much effort is currently being devoted to elucidate whether or not these mutations affect viral transmissibility and virulence. In this review, we summarize the mutations in SARS-CoV-2 during the early phase of virus evolution and discuss the significance of the gene alterations in infections.
Keywords: COVID-19; SARS-CoV-2; bioinformatics; genomes; mutations.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures

References
-
- Tang X, Wu C, Li X, Song Y, Yao X, Wu X, On the origin and continuing evolution of SARS-CoV-2. National Science Review. 2020. https://doi.org/10.1093/nsr/nwaa036. - PMC - PubMed
-
- Control ECfDPa. European Centre for Disease Prevention and Control. 2020. https://www.ecdc.europa.eu/en/geographical-distribution-2019-ncov-cases.
-
- Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020. p:1-4. https://doi.org/10.1038/s41586-020-2012-7. - PMC - PubMed
-
- Xiao K, Zhai J, Feng Y, Zhou N, Zhang X, Zou J-J, Isolation and characterization of 2019-nCoV-like coronavirus from Malayan pangolins. bioRxiv. 2020. https://doi.org/10.1101/2020.02.17.951335.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

