How Population Structure Impacts Genomic Selection Accuracy in Cross-Validation: Implications for Practical Breeding
- PMID: 33391305
- PMCID: PMC7772221
- DOI: 10.3389/fpls.2020.592977
How Population Structure Impacts Genomic Selection Accuracy in Cross-Validation: Implications for Practical Breeding
Abstract
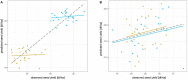
Over the last two decades, the application of genomic selection has been extensively studied in various crop species, and it has become a common practice to report prediction accuracies using cross validation. However, genomic prediction accuracies obtained from random cross validation can be strongly inflated due to population or family structure, a characteristic shared by many breeding populations. An understanding of the effect of population and family structure on prediction accuracy is essential for the successful application of genomic selection in plant breeding programs. The objective of this study was to make this effect and its implications for practical breeding programs comprehensible for breeders and scientists with a limited background in quantitative genetics and genomic selection theory. We, therefore, compared genomic prediction accuracies obtained from different random cross validation approaches and within-family prediction in three different prediction scenarios. We used a highly structured population of 940 Brassica napus hybrids coming from 46 testcross families and two subpopulations. Our demonstrations show how genomic prediction accuracies obtained from among-family predictions in random cross validation and within-family predictions capture different measures of prediction accuracy. While among-family prediction accuracy measures prediction accuracy of both the parent average component and the Mendelian sampling term, within-family prediction only measures how accurately the Mendelian sampling term can be predicted. With this paper we aim to foster a critical approach to different measures of genomic prediction accuracy and a careful analysis of values observed in genomic selection experiments and reported in literature.
Keywords: genomic prediction; nested association mapping population; oilseed rape; predictive breeding; structure.
Copyright © 2020 Werner, Gaynor, Gorjanc, Hickey, Kox, Abbadi, Leckband, Snowdon and Stahl.
Conflict of interest statement
AA and TK were employed by the company NPZ Innovation GmbH. GL was employed by the company German Seed Alliance GmbH. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Clarke E. W., Higgins E. E., Plieske J., Wieseke R., Sidebottom C., Khedikar Y., et al. (2016). A high-density SNP genotyping array for Brassica napus and its ancestral diploid species based on optimised selection of single-locus markers in the allotetraploid genome. Theor. Appl. Genet. 129, 1887–1899. 10.1007/s00122-016-2746-7 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

