Improving the Identification and Coverage of Plant Transmembrane Proteins in Medicago Using Bottom-Up Proteomics
- PMID: 33391307
- PMCID: PMC7775423
- DOI: 10.3389/fpls.2020.595726
Improving the Identification and Coverage of Plant Transmembrane Proteins in Medicago Using Bottom-Up Proteomics
Abstract
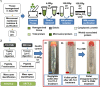
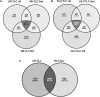
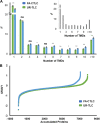
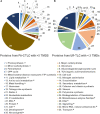
Plant transmembrane proteins (TMPs) are essential for normal cellular homeostasis, nutrient exchange, and responses to environmental cues. Commonly used bottom-up proteomic approaches fail to identify a broad coverage of peptide fragments derived from TMPs. Here, we used mass spectrometry (MS) to compare the effectiveness of two solubilization and protein cleavage methods to identify shoot-derived TMPs from the legume Medicago. We compared a urea solubilization, trypsin Lys-C (UR-TLC) cleavage method to a formic acid solubilization, cyanogen bromide and trypsin Lys-C (FA-CTLC) cleavage method. We assessed the effectiveness of these methods by (i) comparing total protein identifications, (ii) determining how many TMPs were identified, and (iii) defining how many peptides incorporate all, or part, of transmembrane domains (TMD) sequences. The results show that the FA-CTLC method identified nine-fold more TMDs, and enriched more hydrophobic TMPs than the UR-TLC method. FA-CTLC identified more TMPs, particularly transporters, whereas UR-TLC preferentially identified TMPs with one TMD, particularly signaling proteins. The results suggest that combining plant membrane purification techniques with both the FA-CTLC and UR-TLC methods will achieve a more complete identification and coverage of TMPs.
Keywords: Medicago truncatula; TRAMDOMI algorithm; detergent-free purification; liquid chromatography; mass spectrometry; transmembrane domain; transmembrane protein.
Copyright © 2020 Lee, Carroll, Crossett, Connolly, Batarseh and Djordjevic.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Urea free and more efficient sample preparation method for mass spectrometry based protein identification via combining the formic acid-assisted chemical cleavage and trypsin digestion.Talanta. 2011 Oct 30;86:429-35. doi: 10.1016/j.talanta.2011.08.052. Epub 2011 Sep 1. Talanta. 2011. PMID: 22063562
-
Microwave-assisted acid hydrolysis of proteins combined with liquid chromatography MALDI MS/MS for protein identification.J Am Soc Mass Spectrom. 2005 Apr;16(4):471-81. doi: 10.1016/j.jasms.2004.12.017. J Am Soc Mass Spectrom. 2005. PMID: 15792716
-
Structural biology of transmembrane domains: efficient production and characterization of transmembrane peptides by NMR.Protein Sci. 2007 Oct;16(10):2153-65. doi: 10.1110/ps.072996707. Protein Sci. 2007. PMID: 17893361 Free PMC article.
-
Proteomics technologies for the global identification and quantification of proteins.Adv Protein Chem Struct Biol. 2010;80:1-44. doi: 10.1016/B978-0-12-381264-3.00001-1. Adv Protein Chem Struct Biol. 2010. PMID: 21109216 Review.
-
Transmembrane proteins in grape immunity: current knowledge and methodological advances.Front Plant Sci. 2024 Dec 20;15:1515163. doi: 10.3389/fpls.2024.1515163. eCollection 2024. Front Plant Sci. 2024. PMID: 39759230 Free PMC article. Review.
Cited by
-
Evidence That Binding of Cyclic GMP to the Extracellular Domain of NKA (Sodium-Potassium ATPase) Mediates Natriuresis.Circ Res. 2023 Apr 28;132(9):1127-1140. doi: 10.1161/CIRCRESAHA.122.321693. Epub 2023 Mar 15. Circ Res. 2023. PMID: 36919600 Free PMC article.
-
Capillary Zone Electrophoresis-Tandem Mass Spectrometry for Top-Down Proteomics of Mouse Brain Integral Membrane Proteins.Anal Chem. 2023 Aug 29;95(34):12590-12594. doi: 10.1021/acs.analchem.3c02346. Epub 2023 Aug 18. Anal Chem. 2023. PMID: 37595263 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

