Improving the Identification and Coverage of Plant Transmembrane Proteins in Medicago Using Bottom-Up Proteomics
- PMID: 33391307
- PMCID: PMC7775423
- DOI: 10.3389/fpls.2020.595726
Improving the Identification and Coverage of Plant Transmembrane Proteins in Medicago Using Bottom-Up Proteomics
Abstract
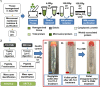
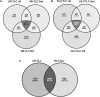
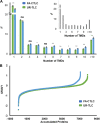
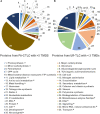
Plant transmembrane proteins (TMPs) are essential for normal cellular homeostasis, nutrient exchange, and responses to environmental cues. Commonly used bottom-up proteomic approaches fail to identify a broad coverage of peptide fragments derived from TMPs. Here, we used mass spectrometry (MS) to compare the effectiveness of two solubilization and protein cleavage methods to identify shoot-derived TMPs from the legume Medicago. We compared a urea solubilization, trypsin Lys-C (UR-TLC) cleavage method to a formic acid solubilization, cyanogen bromide and trypsin Lys-C (FA-CTLC) cleavage method. We assessed the effectiveness of these methods by (i) comparing total protein identifications, (ii) determining how many TMPs were identified, and (iii) defining how many peptides incorporate all, or part, of transmembrane domains (TMD) sequences. The results show that the FA-CTLC method identified nine-fold more TMDs, and enriched more hydrophobic TMPs than the UR-TLC method. FA-CTLC identified more TMPs, particularly transporters, whereas UR-TLC preferentially identified TMPs with one TMD, particularly signaling proteins. The results suggest that combining plant membrane purification techniques with both the FA-CTLC and UR-TLC methods will achieve a more complete identification and coverage of TMPs.
Keywords: Medicago truncatula; TRAMDOMI algorithm; detergent-free purification; liquid chromatography; mass spectrometry; transmembrane domain; transmembrane protein.
Copyright © 2020 Lee, Carroll, Crossett, Connolly, Batarseh and Djordjevic.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

