Ring/U-Box Protein AtUSR1 Functions in Promoting Leaf Senescence Through JA Signaling Pathway in Arabidopsis
- PMID: 33391323
- PMCID: PMC7772223
- DOI: 10.3389/fpls.2020.608589
Ring/U-Box Protein AtUSR1 Functions in Promoting Leaf Senescence Through JA Signaling Pathway in Arabidopsis
Abstract
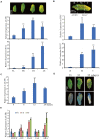
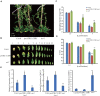
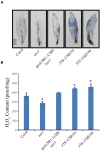
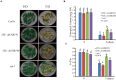
Leaf senescence is regulated by a large number of internal and environmental factors. Here, we report that AtUSR1 (U-box Senescence Related 1) which encodes a plant Ring/U-box protein, is involved in age-dependent and dark-induced leaf senescence in Arabidopsis. Expression of AtUSR1 gene in leaves was up-regulated in darkness and during aging. Plants of usr1, an AtUSR1 gene knock-down mutant, showed a significant delay in age-dependent and dark-induced leaf senescence and the delayed senescence phenotype was rescued when the AtUSR1 gene was transferred back to the mutant plants. Meanwhile, overexpression of AtUSR1 caused accelerated leaf senescence. Furthermore, the role of AtUSR1 in regulating leaf senescence is related to MYC2-mediuated jasmonic acid (JA) signaling pathway. MeJA treatments promoted the accumulation of AtUSR1 transcripts and this expression activation was dependent on the function of MYC2, a key transcription factor in JA signaling. Dual-luciferase assay results indicated that MYC2 promoted the expression of AtUSR1. Overexpression of AtUSR1 in myc2 mutant plants showed precocious senescence, while myc2 mutation alone caused a delay in leaf senescence, suggesting that AtUSR1 functions downstream to MYC2 in the JA signaling pathway in promoting leaf senescence.
Keywords: AtUSR1; JA; MYC2; leaf senescence; ring/U-box.
Copyright © 2020 Zhang, Xu and Guo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Abdollahi M. R., Moieni A., Mousavi A., Salmanian A. H., Jalali Javaran M., Majdi M. (2007). Effect of integrated bombardment and Agrobacterium transformation system on transient GUS expression in hypocotyls of rapeseed (Brassica napus L. cv. PF704) microspore-derived embryos. Pak. J. Biol. Sci. 10 3141–3145. 10.3923/pjbs.2007.3141.3145 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

