Performance of at-home self-collected saliva and nasal-oropharyngeal swabs in the surveillance of COVID-19
- PMID: 33391631
- PMCID: PMC7733974
- DOI: 10.1080/20002297.2020.1858002
Performance of at-home self-collected saliva and nasal-oropharyngeal swabs in the surveillance of COVID-19
Abstract
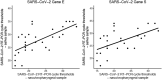
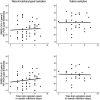
Background: SARS-CoV-2 quickly spreads in the worldwide population, imposing social restrictions to control the infection, being the massive testing another essential strategy to break the chain of transmission. Aim: To compare the performance of at-home self-collected samples - saliva and combined nasal-oropharyngeal swabs (NOP) - for SARS-CoV-2 detection in a telemedicine platform for COVID-19 surveillance. Material and methods: We analyzed 201 patients who met the criteria of suspected COVID-19. NOP sampling was combined (nostrils and oropharynx) and saliva collected using a cotton pad device. Detection of SARS-COV-2 was performed by using the Altona RealStar® SARS-CoV-2 RT-PCR Kit 1.0. Results: There was an overall significant agreement (κ coefficient value of 0.58) between saliva and NOP. Considering results in either sample, 70 patients positive for SARS-CoV-2 were identified, with 52/70 being positive in NOP and 55/70 in saliva. This corresponds to sensitivities of 74.2% (95% CI; 63.7% to 83.1%) for NOP and 78.6% (95% CI; 67.6% to 86.6%) for saliva. Conclusion: Our data show the feasibility of using at-home self-collected samples (especially saliva), as an adequate alternative for SARS-CoV-2 detection. This new approach of testing can be useful to develop strategies for COVID-19 surveillance and for guiding public health decisions.
Keywords: PCR; Saliva; coronavirus; infection control; primary health care; telemedicine.
© 2020 The Author(s). Published by Informa UK Limited, trading as Taylor & Francis Group.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures

References
-
- Ott IM, Strine MS, Watkins AE, et al. Simply saliva: stability of SARS-CoV-2 detection negates the need for expensive collection devices. medRxiv. 2020. DOI:10.1101/2020.08.03.20165233 - DOI
-
- Valentine-Graves M, Hall E, Guest JL, et al. At-home self-collection of saliva, oropharyngeal swabs and dried blood spots for SARS-CoV-2 diagnosis and serology: post-collection acceptability of specimen collection process and patient confidence in specimens. PLoS One. 2020;15:e0236775. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
