Identification of key genes involved in post-traumatic stress disorder: Evidence from bioinformatics analysis
- PMID: 33392005
- PMCID: PMC7754529
- DOI: 10.5498/wjp.v10.i12.286
Identification of key genes involved in post-traumatic stress disorder: Evidence from bioinformatics analysis
Abstract
Background: Post-traumatic stress disorder (PTSD) is a serious stress-related disorder.
Aim: To identify the key genes and pathways to uncover the potential mechanisms of PTSD using bioinformatics methods.
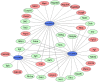
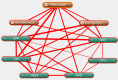
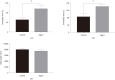
Methods: Gene expression profiles were obtained from the Gene Expression Omnibus database. The differentially expressed genes (DEGs) were identified by using GEO2R. Gene functional annotation and pathway enrichment were then conducted. The gene-pathway network was constructed with Cytoscape software. Quantitative real-time polymerase chain reaction (qRT-PCR) analysis was applied for validation, and text mining by Coremine Medical was used to confirm the connections among genes and pathways.
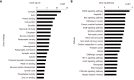
Results: We identified 973 DEGs including 358 upregulated genes and 615 downregulated genes in PTSD. A group of centrality hub genes and significantly enriched pathways (MAPK, Ras, and ErbB signaling pathways) were identified by using gene functional assignment and enrichment analyses. Six genes (KRAS, EGFR, NFKB1, FGF12, PRKCA, and RAF1) were selected to validate using qRT-PCR. The results of text mining further confirmed the correlation among hub genes and the enriched pathways. It indicated that these altered genes displayed functional roles in PTSD via these pathways, which might serve as key signatures in the pathogenesis of PTSD.
Conclusion: The current study identified a panel of candidate genes and important pathways, which might help us deepen our understanding of the underlying mechanism of PTSD at the molecular level. However, further studies are warranted to discover the critical regulatory mechanism of these genes via relevant pathways in PTSD.
Keywords: Bioinformatics analysis; Differentially expressed genes; Gene-pathway co-expression; Key pathway; Microarray; Post-traumatic stress disorder.
©The Author(s) 2020. Published by Baishideng Publishing Group Inc. All rights reserved.
Conflict of interest statement
Conflict-of-interest statement: The authors declare no conflicts of interest.
Figures


Similar articles
-
Identification of Key Genes and Pathways in Post-traumatic Stress Disorder Using Microarray Analysis.Front Psychol. 2019 Feb 27;10:302. doi: 10.3389/fpsyg.2019.00302. eCollection 2019. Front Psychol. 2019. PMID: 30873067 Free PMC article.
-
Identification of Critical Signature in Post-Traumatic Stress Disorder Using Bioinformatics Analysis and in Vitro Analyses.Brain Behav. 2025 Jan;15(1):e70243. doi: 10.1002/brb3.70243. Brain Behav. 2025. PMID: 39829117 Free PMC article.
-
The identification of key genes and pathways in hepatocellular carcinoma by bioinformatics analysis of high-throughput data.Med Oncol. 2017 Jun;34(6):101. doi: 10.1007/s12032-017-0963-9. Epub 2017 Apr 21. Med Oncol. 2017. PMID: 28432618 Free PMC article.
-
Bioinformatics analyses of significant genes, related pathways and candidate prognostic biomarkers in glioblastoma.Mol Med Rep. 2018 Nov;18(5):4185-4196. doi: 10.3892/mmr.2018.9411. Epub 2018 Aug 21. Mol Med Rep. 2018. PMID: 30132538 Free PMC article.
-
Identification of candidate biomarkers and pathways associated with SCLC by bioinformatics analysis.Mol Med Rep. 2018 Aug;18(2):1538-1550. doi: 10.3892/mmr.2018.9095. Epub 2018 May 29. Mol Med Rep. 2018. PMID: 29845250 Free PMC article.
Cited by
-
Using Monozygotic Twins to Dissect Common Genes in Posttraumatic Stress Disorder and Migraine.Front Neurosci. 2021 Jun 22;15:678350. doi: 10.3389/fnins.2021.678350. eCollection 2021. Front Neurosci. 2021. PMID: 34239411 Free PMC article.
-
The relationship between social adversity, micro-RNA expression and post-traumatic stress in a prospective, community-based cohort.Res Sq [Preprint]. 2025 Mar 17:rs.3.rs-5867503. doi: 10.21203/rs.3.rs-5867503/v1. Res Sq. 2025. PMID: 40166034 Free PMC article. Preprint.
References
-
- Paredes Molina CS, Berry S, Nielsen A, Winfield R. PTSD in civilian populations after hospitalization following traumatic injury: A comprehensive review. Am J Surg. 2018;216:745–753. - PubMed
-
- Qi W, Ratanatharathorn A, Gevonden M, Bryant R, Delahanty D, Matsuoka Y, Olff M, deRoon-Cassini T, Schnyder U, Seedat S, Laska E, Kessler RC, Koenen K, Shalev A on behalf of the ICPP. Application of data pooling to longitudinal studies of early post-traumatic stress disorder (PTSD): the International Consortium to Predict PTSD (ICPP) project. Eur J Psychotraumatol. 2018;9:1476442. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

