Near haploidization is a genomic hallmark which defines a molecular subgroup of giant cell glioblastoma
- PMID: 33392505
- PMCID: PMC7764500
- DOI: 10.1093/noajnl/vdaa155
Near haploidization is a genomic hallmark which defines a molecular subgroup of giant cell glioblastoma
Abstract
Background: Giant cell glioblastoma (gcGBM) is a rare histologic subtype of glioblastoma characterized by numerous bizarre multinucleate giant cells and increased reticulin deposition. Compared with conventional isocitrate dehydrogenase (IDH)-wildtype glioblastomas, gcGBMs typically occur in younger patients and are generally associated with an improved prognosis. Although prior studies of gcGBMs have shown enrichment of genetic events, such as TP53 alterations, no defining aberrations have been identified. The aim of this study was to evaluate the genomic profile of gcGBMs to facilitate more accurate diagnosis and prognostication for this entity.
Methods: Through a multi-institutional collaborative effort, we characterized 10 gcGBMs by chromosome studies, single nucleotide polymorphism microarray analysis, and targeted next-generation sequencing. These tumors were subsequently compared to the genomic and epigenomic profile of glioblastomas described in The Cancer Genome Atlas (TCGA) dataset.
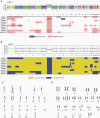
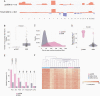
Results: Our analysis identified a specific pattern of genome-wide massive loss of heterozygosity (LOH) driven by near haploidization in a subset of glioblastomas with giant cell histology. We compared the genomic signature of these tumors against that of all glioblastomas in the TCGA dataset (n = 367) and confirmed that our cohort of gcGBMs demonstrated a significantly different genomic profile. Integrated genomic and histologic review of the TCGA cohort identified 3 additional gcGBMs with a near haploid genomic profile.
Conclusions: Massive LOH driven by haploidization represents a defining molecular hallmark of a subtype of gcGBM. This unusual mechanism of tumorigenesis provides a diagnostic genomic hallmark to evaluate in future cases, may explain reported differences in survival, and suggests new therapeutic vulnerabilities.
Keywords: giant cell glioblastoma; glioblastoma; loss of heterozygosity; near haploidization.
© The Author(s) 2020. Published by Oxford University Press, the Society for Neuro-Oncology and the European Association of Neuro-Oncology.
Figures

References
-
- Louis DN, Ohgaki H, Wiestler OD, et al., eds. WHO Classification of Tumours of the Central Nervous System. Revised 4th ed. Lyon, France: International Agency for Research on Cancer; 2016.
-
- Ortega A, Nuño M, Walia S, Mukherjee D, Black KL, Patil CG. Treatment and survival of patients harboring histological variants of glioblastoma. J Clin Neurosci. 2014;21(10):1709–1713. - PubMed
-
- Jin MC, Wu A, Xiang M, et al. . Prognostic factors and treatment patterns in the management of giant cell glioblastoma. World Neurosurg. 2019;128:e217–e224. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
