Comparative RNA-Seq analysis reveals genes associated with masculinization in female Cannabis sativa
- PMID: 33392743
- PMCID: PMC7779414
- DOI: 10.1007/s00425-020-03522-y
Comparative RNA-Seq analysis reveals genes associated with masculinization in female Cannabis sativa
Abstract
Using RNA profiling, we identified several silver thiosulfate-induced genes that potentially control the masculinization of female Cannabis sativa plants. Genetically female Cannabis sativa plants normally bear female flowers, but can develop male flowers in response to environmental and developmental cues. In an attempt to elucidate the molecular elements responsible for sex expression in C. sativa plants, we developed genetically female lines producing both female and chemically-induced male flowers. Furthermore, we carried out RNA-Seq assays aimed at identifying differentially expressed genes responsible for male flower development in female plants. The results revealed over 10,500 differentially expressed genes, of which around 200 potentially control masculinization of female cannabis plants. These genes include transcription factors and other genes involved in male organ (i.e., anther and pollen) development, as well as genes involved in phytohormone signalling and male-biased phenotypes. The expressions of 15 of these genes were further validated by qPCR assay confirming similar expression patterns to that of RNA-Seq data. These genes would be useful for understanding predisposed plants producing flowers of both sex types in the same plant, and help breeders to regulate the masculinization of female plants through targeted breeding and plant biotechnology.
Keywords: Cannabis; Differential expression; Flower sex; Male biased-genes; RNA-seq; Silver thiosulfate.
Figures

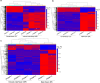
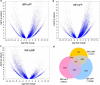

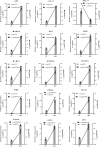
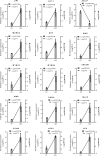
References
-
- Bernstein N, Gorelick J, Koch S. Interplay between chemistry and morphology in medical cannabis (Cannabis sativa L.) Ind Crops Prod. 2019;129:185–194. doi: 10.1016/j.indcrop.2018.11.039. - DOI
-
- Borthwick HA, Scully NJ (1954) Photoperiodic Responses of Hemp. Botanical gazette, 116:14–29. https://www.jstor.org/stable/2473219
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

