Actin filament oxidation by MICAL1 suppresses protections from cofilin-induced disassembly
- PMID: 33393173
- PMCID: PMC7857426
- DOI: 10.15252/embr.202050965
Actin filament oxidation by MICAL1 suppresses protections from cofilin-induced disassembly
Abstract
Proteins of the ADF/cofilin family play a central role in the disassembly of actin filaments, and their activity must be tightly regulated in cells. Recently, the oxidation of actin filaments by the enzyme MICAL1 was found to amplify the severing action of cofilin through unclear mechanisms. Using single filament experiments in vitro, we found that actin filament oxidation by MICAL1 increases, by several orders of magnitude, both cofilin binding and severing rates, explaining the dramatic synergy between oxidation and cofilin for filament disassembly. Remarkably, we found that actin oxidation bypasses the need for cofilin activation by dephosphorylation. Indeed, non-activated, phosphomimetic S3D-cofilin binds and severs oxidized actin filaments rapidly, in conditions where non-oxidized filaments are unaffected. Finally, tropomyosin Tpm1.8 loses its ability to protect filaments from cofilin severing activity when actin is oxidized by MICAL1. Together, our results show that MICAL1-induced oxidation of actin filaments suppresses their physiological protection from the action of cofilin. We propose that, in cells, direct post-translational modification of actin filaments by oxidation is a way to trigger their disassembly.
Keywords: ADF/cofilin; actin dynamics; actin-binding proteins; microfluidics.
© 2021 The Authors.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

- A
Three steps of a typical experiment (see also Materials and Methods). Filaments are polymerized with 0.6–1 µM ATP‐G‐actin and aged for 15 min with ATP‐G‐actin at critical concentration (0.1 µM) to maintain the filament length. This solution is supplemented with MICAL1 and NADPH to oxidize filaments. Tpm can also be added at this step to fully decorate filaments (Figs 3 and 4).
- B
Fraction (1/17th) of a typical field of view. In a microfluidic chamber, actin filaments (yellow) are anchored by their pointed ends and align with the flow.
- C
Time‐lapse images showing the assembly of cofilin‐1 domains (blue) and subsequent severing of actin filaments (yellow). The time‐lapse of an oxidized filament shows filament severing at 6 s; the first cofilin domain nucleation at 2 s; growth of four individual domains as their fluorescence intensity increases; and severing (lightning symbol) at the top domain, 2 s after its nucleation.
- D
Global measurement of the severing of filaments exposed to 20 nM mCherry‐cofilin‐1 from time t = 0 onwards. N = 20 (from one representative experiment) and 50 (1 experiment) for non‐oxidized and oxidized actin filaments, respectively. P‐value = 8 × 10−8 (log‐rank test).
- E
Nucleation of the first cofilin domain onto 5 µm long actin segments. Filaments are exposed to 20 nM mCherry‐cofilin‐1 from time t = 0 onwards. N = 60 filaments, from one experiment for each condition. P‐value = 4 × 10−15 (log‐rank test).
- F
Growth rate of single cofilin domains, normalized by the cofilin concentration. N = 50 (from three independent experiments) and 20 (1 experiment) domains on non‐oxidized and oxidized filaments, respectively. Measurements were obtained using 20 and 100 nM cofilin (non‐oxidized actin) and 10 nM cofilin (oxidized actin). Note that this normalized growth rate does not depend on the cofilin concentration (Appendix Fig S4A). Bars: mean and SD. P‐value = 7 × 10−8 (t‐test).
- G
Filament severing rate at single cofilin domains. Time t = 0 is defined for every domain as the frame on which they nucleate. N = 163 (from two independent experiments) and 203 (3 experiments) cofilin domains on non‐oxidized and oxidized actin filaments, respectively. Measurements were performed at 100 nM cofilin (non‐oxidized F‐actin) and 10, 20, or 30 nM cofilin (oxidized F‐actin). Since the severing rate does not depend on the cofilin concentration (Appendix Fig S4B and (Wioland et al, 2017, 2019a)), data for 10, 20, and 30 nM cofilin on oxidized filaments were pooled. Measurements on non‐oxidized filaments were done at 100 nM mCherry‐cofilin‐1. P‐value = 9 × 10−41 (log‐rank test).
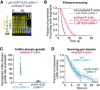
- A
Time‐lapse images of eGFP‐S3D‐cofilin‐1 (blue) binding and severing an oxidized actin filament (yellow).
- B
Global quantification of the severing events on oxidized and non‐oxidized filaments exposed to unlabeled S3D‐cofilin‐1 from time t = 0 onwards. From top to bottom, N = 50 (from 1 experiment), 50 (1 experiment), 100 (2 experiments) filaments.
- C
Growth rate of single cofilin domains, normalized by the cofilin concentration. N = 20 domains for WT mCherry‐cofilin‐1 (1 experiment) and eGFP‐S3D‐cofilin‐1 (2 experiments). The growth rate was measured at 10 nM WT cofilin‐1, and 500 nM and 1 µM S3D‐cofilin‐1 (Appendix Fig S4C). Bars: mean and SD. P‐value = 8 × 10−9 (t‐test).
- D
Filament severing at single cofilin domains. Time t = 0 is defined for every domain as the frame on which it nucleates. N = 60 (1 experiment) and 75 (2 experiments) domains of WT and S3D‐cofilin‐1, respectively. Experiments were performed at 10 nM WT eGFP‐cofilin‐1 and 1 µM eGFP‐S3D‐cofilin‐1. P‐value = 0.98 (log‐rank test).

- A
Time‐lapse images of Tpm‐saturated actin filaments depolymerizing from their barbed end (BE), with or without MICAL1. Depolymerization is initiated at time t = 0 from a non‐oxidized actin filament. The increase in the depolymerization rate reflects the oxidation of actin with time. Tpm is labeled with neonGreen (nGr), and actin is unlabeled.
- B
Barbed end depolymerization rate as a function of the exposure time to either 100 nM nGr‐Tpm1.8 alone (“no MICAL1”) or supplemented with 100 nM MICAL1 and 12 µM NADPH. N = 31 (1 experiment) and 29 (1 experiment) filaments, without and with MICAL1, respectively. Lines: mean velocity. Shaded surfaces: S.D.
- C
Time‐lapse images showing the disassembly of Tpm domains. Filaments were saturated with 100 nM nGr‐Tpm1.8 before the experiment and Tpm was removed from solution at time t = 0 (the solution contains 0.6 µM unlabeled G‐actin to prevent filament depolymerization). PE and BE indicate F‐actin pointed and barbed end, respectively.
- D
Disassembly rate of single Tpm domains, at boundaries located toward the pointed end (PE) and toward the barbed end (BE) of actin filaments. N = 100 (non‐oxidized F‐actin, four experiments) and 50 (oxidized F‐actin, two experiments) domains per direction. Bars: mean and S.D. P‐value (t‐test, comparing oxidized and non‐oxidized filaments) = 0.27 (BE) and 0.12 (PE).
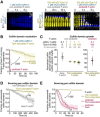
- A
Time‐lapse images of filaments, oxidized or not, bare or Tpm‐saturated, exposed to 1 µM mCherry‐cofilin‐1, from time t = 0 onwards. Other examples are shown in Appendix Fig S6.
- B
Nucleation of the first cofilin domain onto 5 µm long Tpm‐saturated actin filaments. Filaments are exposed to 1 µM mCherry‐cofilin from time t = 0 onwards. N = 50 filaments from one experiment for each condition. P‐value = 4× 10−15 (log‐rank test).
- C
Growth rate of single cofilin domains, normalized by the cofilin concentration. Filaments saturated by Tpm during aging (Fig 1A) were then exposed to cofilin, along with 0–100 nM nGr‐Tpm1.8 to test the competition between the two soluble proteins. Note the scale difference for oxidized filaments. Bars: Mean and S.D. Number of experiments, first to last condition: 2, 1, 1, 3, and 1.
- D, E
Filament severing rate at single cofilin domains. Time t = 0 is defined for every domain as the frame on which they nucleate. N = 163 (2 experiments), 180 (4 experiments), 116 (3 experiments), 203 (3 experiments) domains for bare actin (100 nM mCherry‐cofilin‐1), non‐oxidized Tpm‐saturated (1 µM mCherry‐cofilin‐1), oxidized Tpm‐saturated filaments (100 nM mCherry‐cofilin‐1), and bare oxidized actin (10, 20, 30 nM mCherry‐cofilin‐1), respectively.

- A
Time‐lapse of actin filaments constantly exposed to nGr‐Tpm1.8 (yellow), WT mCh‐cofilin‐1 (blue) and S3D‐cofilin‐1, and to MICAL1 from time t = 100 s onwards. Here we mixed WT with an excess of S3D‐cofilin‐1 to mimic cellular conditions in which most of cofilin‐1 is inhibited by phosphorylation. While Tpm prevents cofilin from binding to non‐oxidized filaments, MICAL1 rapidly oxidizes actin, allowing cofilin to bind and sever filaments. See also Supp Movie.
- B
Summary of the results. Different mechanisms can simultaneously down regulate the activity of cofilin (here cofilin phosphorylation and protection by Tpm). Actin oxidation is a rapid means to cancel these protections without, for example, the need to activate a large concentration of cofilin.
References
-
- Arber S, Barbayannis FA, Hanser H, Schneider C, Stanyon CA, Bernard O, Caroni P (1998) Regulation of actin dynamics through phosphorylation of cofilin by LIM‐kinase. Nature 393: 805–809 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

