Two-stage genome-wide association study of chronic rhinosinusitis and disease subphenotypes highlights mucosal immunity contributing to risk
- PMID: 33393196
- PMCID: PMC8048969
- DOI: 10.1002/alr.22731
Two-stage genome-wide association study of chronic rhinosinusitis and disease subphenotypes highlights mucosal immunity contributing to risk
Keywords: Chronic Rhinosinusitis; Rhinosinusitis; Statistics.
Figures
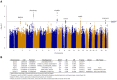
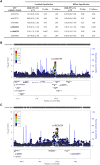
References
-
- Kristjansson RP, Benonisdottir S, Davidsson OB, et al. A loss‐of‐function variant in ALOX15 protects against nasal polyps and chronic rhinosinusitis. Nat Genet. 2019;51:267‐276. - PubMed
-
- Fokkens WJ, Lund VJ, Mullol J, et al. EPOS 2012: European Position Paper on Rhinosinusitis and Nasal Polyps 2012. A summary for otorhinolaryngologists. Rhinology. 2012;50:1‐12. - PubMed
-
- Muthén LK, Muthén BO. Mplus User's Guide: Statistical Analysis with Latent Variables. 7th ed. Los Angeles, CA: Muthén & Muthén; 1998‐2012.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

