TLR3 controls constitutive IFN-β antiviral immunity in human fibroblasts and cortical neurons
- PMID: 33393505
- PMCID: PMC7773389
- DOI: 10.1172/JCI134529
TLR3 controls constitutive IFN-β antiviral immunity in human fibroblasts and cortical neurons
Abstract
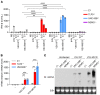
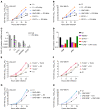
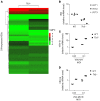
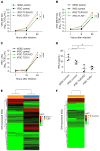
Human herpes simplex virus 1 (HSV-1) encephalitis can be caused by inborn errors of the TLR3 pathway, resulting in impairment of CNS cell-intrinsic antiviral immunity. Deficiencies of the TLR3 pathway impair cell-intrinsic immunity to vesicular stomatitis virus (VSV) and HSV-1 in fibroblasts, and to HSV-1 in cortical but not trigeminal neurons. The underlying molecular mechanism is thought to involve impaired IFN-α/β induction by the TLR3 recognition of dsRNA viral intermediates or by-products. However, we show here that human TLR3 controls constitutive levels of IFNB mRNA and secreted bioactive IFN-β protein, and thereby also controls constitutive mRNA levels for IFN-stimulated genes (ISGs) in fibroblasts. Tlr3-/- mouse embryonic fibroblasts also have lower basal ISG levels. Moreover, human TLR3 controls basal levels of IFN-β secretion and ISG mRNA in induced pluripotent stem cell-derived cortical neurons. Consistently, TLR3-deficient human fibroblasts and cortical neurons are vulnerable not only to both VSV and HSV-1, but also to several other families of viruses. The mechanism by which TLR3 restricts viral growth in human fibroblasts and cortical neurons in vitro and, by inference, by which the human CNS prevents infection by HSV-1 in vivo, is therefore based on the control of early viral infection by basal IFN-β immunity.
Keywords: Immunology; Infectious disease; Innate immunity.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

