Toward an Information Theory of Quantitative Genetics
- PMID: 33395537
- PMCID: PMC8220575
- DOI: 10.1089/cmb.2020.0032
Toward an Information Theory of Quantitative Genetics
Abstract
Quantitative genetics has evolved dramatically in the past century, and the proliferation of genetic data, in quantity as well as type, enables the characterization of complex interactions and mechanisms beyond the scope of its theoretical foundations. In this article, we argue that revisiting the framework for analysis is important and we begin to lay the foundations of an alternative formulation of quantitative genetics based on information theory. Information theory can provide sensitive and unbiased measures of statistical dependencies among variables, and it provides a natural mathematical language for an alternative view of quantitative genetics. In the previous work, we examined the information content of discrete functions and applied this approach and methods to the analysis of genetic data. In this article, we present a framework built around a set of relationships that both unifies the information measures for the discrete functions and uses them to express key quantitative genetic relationships. Information theory measures of variable interdependency are used to identify significant interactions, and a general approach is described for inferring functional relationships in genotype and phenotype data. We present information-based measures of the genetic quantities: penetrance, heritability, and degrees of statistical epistasis. Our scope here includes the consideration of both two- and three-variable dependencies and independently segregating variants, which captures additive effects, genetic interactions, and two-phenotype pleiotropy. This formalism and the theoretical approach naturally apply to higher multivariable interactions and complex dependencies, and can be adapted to account for population structure, linkage, and nonrandomly segregating markers. This article thus focuses on presenting the initial groundwork for a full formulation of quantitative genetics based on information theory.
Keywords: entropy; epistasis; genetics; information theory.
Conflict of interest statement
The authors declare they have no competing financial interests.
Figures
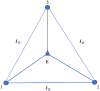
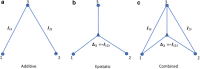
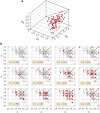
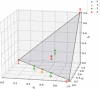

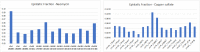
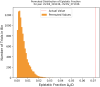
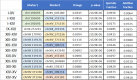

References
-
- Bell, A.J. 2003. The co-information lattice. In ICA 2003, Nara, Japan, 921–926
-
- Bertschinger, N., Rauh, J., Olbrich, E., et al. 2012. Shared Information: New insights and problems in decomposing information in complex systems, 251–269. Proceedings of the ECCS. Springer, Cham
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

