Unpacking Pandora From Its Box: Deciphering the Molecular Basis of the SARS-CoV-2 Coronavirus
- PMID: 33396557
- PMCID: PMC7795774
- DOI: 10.3390/ijms22010386
Unpacking Pandora From Its Box: Deciphering the Molecular Basis of the SARS-CoV-2 Coronavirus
Abstract
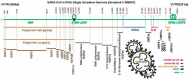
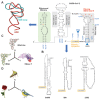
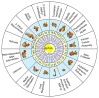
An enigmatic localized pneumonia escalated into a worldwide COVID-19 pandemic from Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). This review aims to consolidate the extensive biological minutiae of SARS-CoV-2 which requires decipherment. Having one of the largest RNA viral genomes, the single strand contains the genes ORF1ab, S, E, M, N and ten open reading frames. Highlighting unique features such as stem-loop formation, slippery frameshifting sequences and ribosomal mimicry, SARS-CoV-2 represents a formidable cellular invader. Hijacking the hosts translational engine, it produces two polyprotein repositories (pp1a and pp1ab), armed with self-cleavage capacity for production of sixteen non-structural proteins. Novel glycosylation sites on the spike trimer reveal unique SARS-CoV-2 features for shielding and cellular internalization. Affording complexity for superior fitness and camouflage, SARS-CoV-2 challenges diagnosis and vaccine vigilance. This review serves the scientific community seeking in-depth molecular details when designing drugs to curb transmission of this biological armament.
Keywords: 2019-nCoV; COVID-19; RNA; bats; coronavirus; pandemic; virus.
Conflict of interest statement
All authors declare no conflict of interest.
Figures
Similar articles
-
[Basic information of Coronavirus].Uirusu. 2020;70(1):29-36. doi: 10.2222/jsv.70.29. Uirusu. 2020. PMID: 33967109 Review. Japanese.
-
SARS-CoV-2 strategically mimics proteolytic activation of human ENaC.Elife. 2020 May 26;9:e58603. doi: 10.7554/eLife.58603. Elife. 2020. PMID: 32452762 Free PMC article.
-
Latent periodicity-2 in coronavirus SARS-CoV-2 genome: Evolutionary implications.J Theor Biol. 2021 Apr 21;515:110604. doi: 10.1016/j.jtbi.2021.110604. Epub 2021 Jan 26. J Theor Biol. 2021. PMID: 33508323 Free PMC article.
-
The coding capacity of SARS-CoV-2.Nature. 2021 Jan;589(7840):125-130. doi: 10.1038/s41586-020-2739-1. Epub 2020 Sep 9. Nature. 2021. PMID: 32906143
-
Properties of Coronavirus and SARS-CoV-2.Malays J Pathol. 2020 Apr;42(1):3-11. Malays J Pathol. 2020. PMID: 32342926 Review.
Cited by
-
Drug Repurposing for COVID-19: A Review and a Novel Strategy to Identify New Targets and Potential Drug Candidates.Molecules. 2022 Apr 23;27(9):2723. doi: 10.3390/molecules27092723. Molecules. 2022. PMID: 35566073 Free PMC article. Review.
-
Viral-Induced Inflammatory Coagulation Disorders: Preparing for Another Epidemic.Thromb Haemost. 2022 Jan;122(1):8-19. doi: 10.1055/a-1562-7599. Epub 2021 Sep 13. Thromb Haemost. 2022. PMID: 34331297 Free PMC article. Review.
-
Sec61 Inhibitor Apratoxin S4 Potently Inhibits SARS-CoV-2 and Exhibits Broad-Spectrum Antiviral Activity.ACS Infect Dis. 2022 Jul 8;8(7):1265-1279. doi: 10.1021/acsinfecdis.2c00008. Epub 2022 Jun 29. ACS Infect Dis. 2022. PMID: 35766385 Free PMC article.
References
-
- Gates B. [Video File]. Ted Talk2015 The Next Outbreak? We’re Not Ready. [(accessed on 29 December 2020)]; Available online: https://www.ted.com/talks/bill_gates_the_next_outbreak_we_re_not_ready?l....
-
- Commission WMH . Report of Clustering Pneumonia of Unknown Etiology in Wuhan City. Commission WMH; Wuhan, China: 2020.
-
- Lai C.-C., Wang C.-Y., Wang Y.-H., Hsueh S.-C., Ko W.-C., Hsueh P.-R. Global epidemiology of coronavirus disease 2019 (COVID-19): Disease incidence, daily cumulative index, mortality, and their association with country healthcare resources and economic status. Int. J. Antimicrob. Agents. 2020;55:105946. doi: 10.1016/j.ijantimicag.2020.105946. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

