Histone methyltransferase SUV39H2 regulates LSD1-dependent CDH1 expression and promotes epithelial mesenchymal transition of osteosarcoma
- PMID: 33397384
- PMCID: PMC7784292
- DOI: 10.1186/s12935-020-01636-7
Histone methyltransferase SUV39H2 regulates LSD1-dependent CDH1 expression and promotes epithelial mesenchymal transition of osteosarcoma
Retraction in
-
Retraction Note: Histone methyltransferase SUV39H2 regulates LSD1-dependent CDH1 expression and promotes epithelial mesenchymal transition of osteosarcoma.Cancer Cell Int. 2023 Aug 18;23(1):173. doi: 10.1186/s12935-023-03016-3. Cancer Cell Int. 2023. PMID: 37596581 Free PMC article. No abstract available.
Abstract
Objective: Osteosarcoma (OS) is a malignant tumor characterized by the direct production of bone or osteoid tissues by proliferating tumor cells. Suppressor of variegation 3-9 homolog 2 (SUV39H2) is implicated in the occurrence of OS. Therefore, we designed this study to investigate effects of SUV39H2 in OS meditated by the lysine specific demethylase-1/E-cadherin (LSD1/CDH1) axis.
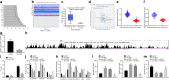
Methods: Clinical OS tissues and paracancerous tissues were collected for analysis of SUV39H2, LSD1 and CDH1 expression, and Kaplan-Meier survival analysis was applied to test the relationship between SUV39H2 expression and overall survival. Loss- and gain-of-function assays were conducted to determine the roles of SUV39H2, LSD1 and CDH1 in OS epithelial mesenchymal transition (EMT) and migration in OS cells, with quantitation of relevant proteins by immunofluorescence. We confirmed the effects of modulating the SUV39H2/CDH1 axis in a mouse OS tumor model.
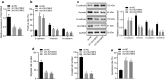
Results: SUV39H2 and LSD1 were highly expressed, while CDH1 was downregulated in OS tissues and cells. SUV39H2 expression correlated inversely with overall survival of patients with OS. SUV39H2 positively regulated LSD1 expression, while LSD1 negatively regulated CDH1 expression. SUV39H2 or LSD1 overexpression, or CDH1 silencing promoted migration and EMT, as indicated by reduced E-cadherin and dramatically upregulated Vimentin and N-cadherin of OS cells. SUV39H2 expedited the progression of OS, which was reversed by CDH1 repression in the setting of OS in vitro and in vivo.
Conclusions: Collectively, our results demonstrate highly expressed SUV39H2 in OS elevates the expression of LSD1 to downregulate CDH1 expression, thereby aggravating OS, providing a potential therapeutic target for treatment of OS.
Keywords: E-cadherin gene; Histone methyltransferases; Lysine specific demethylase-1; Osteosarcoma; Suppressor of variegation 3–9 homolog 2.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


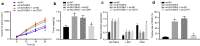
References
-
- Tian H, Zhou T, Chen H, Li C, Jiang Z, Lao L, Kahn SA, Duarte MEL, Zhao J, Daubs MD, et al. Bone morphogenetic protein-2 promotes osteosarcoma growth by promoting epithelial-mesenchymal transition (EMT) through the Wnt/beta-catenin signaling pathway. J Orthop Res. 2019;37(7):1638–48. doi: 10.1002/jor.24244. - DOI - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

