The Operon Encoding Hydrolytic Dehalogenation of 4-Chlorobenzoate Is Transcriptionally Regulated by the TetR-Type Repressor FcbR and Its Ligand 4-Chlorobenzoyl Coenzyme A
- PMID: 33397703
- PMCID: PMC8105013
- DOI: 10.1128/AEM.02652-20
The Operon Encoding Hydrolytic Dehalogenation of 4-Chlorobenzoate Is Transcriptionally Regulated by the TetR-Type Repressor FcbR and Its Ligand 4-Chlorobenzoyl Coenzyme A
Abstract
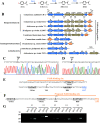
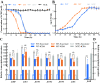
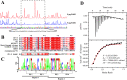
The bacterial hydrolytic dehalogenation of 4-chlorobenzoate (4CBA) is a coenzyme A (CoA)-activation-type catabolic pathway that is usually a common part of the microbial mineralization of chlorinated aromatic compounds. Previous studies have shown that the transport and dehalogenation genes for 4CBA are typically clustered as an fcbBAT1T2T3C operon and inducibly expressed in response to 4CBA. However, the associated molecular mechanism remains unknown. In this study, a gene (fcbR) adjacent to the fcb operon was predicted to encode a TetR-type transcriptional regulator in Comamonas sediminis strain CD-2. The fcbR knockout strain exhibited constitutive expression of the fcb cluster. In the host Escherichia coli, the expression of the Pfcb -fused green fluorescent protein (gfp) reporter was repressed by the introduction of the fcbR gene, and genetic studies combining various catabolic genes suggest that the ligand for FcbR may be an intermediate metabolite. Purified FcbR could bind to the Pfcb DNA probe in vitro, and the metabolite 4-chlorobenzyl-CoA (4CBA-CoA) prevented FcbR binding to the P fcb DNA probe. Isothermal titration calorimetry (ITC) measurements showed that 4CBA-CoA could bind to FcbR at a 1:1 molar ratio. DNase I footprinting showed that FcbR protected a 42-bp DNA motif (5'-GGAAATCAATAGGTCCATAGAAAATCTATTGACTAATCGAAT-3') that consists of two sequence repeats containing four pseudopalindromic sequences (5'-TCNATNGA-3'). This binding motif overlaps with the -35 box of Pfcb and was proposed to prevent the binding of RNA polymerase. This study characterizes a transcriptional repressor of the fcb operon, together with its ligand, thus identifying halogenated benzoyl-CoA as belonging to the class of ligands of transcriptional regulators.IMPORTANCE The bacterial hydrolytic dehalogenation of 4CBA is a special CoA-activation-type catabolic pathway that plays an important role in the biodegradation of polychlorinated biphenyls and some herbicides. With genetic and biochemical approaches, the present study identified the transcriptional repressor and its cognate effector of a 4CBA hydrolytic dehalogenation operon. This work extends halogenated benzoyl-CoA as a new member of CoA-derived effector compounds that mediate allosteric regulation of transcriptional regulators.
Keywords: 4-chlorobenzoate; 4-chlorobenzoyl-CoA; FcbR; TetR-type transcriptional regulator; hydrolytic dehalogenation.
Copyright © 2021 American Society for Microbiology.
Figures



Similar articles
-
Phenylacetyl coenzyme A is an effector molecule of the TetR family transcriptional repressor PaaR from Thermus thermophilus HB8.J Bacteriol. 2011 Sep;193(17):4388-95. doi: 10.1128/JB.05203-11. Epub 2011 Jul 1. J Bacteriol. 2011. PMID: 21725002 Free PMC article.
-
4-Chlorobenzoate uptake in Comamonas sp. strain DJ-12 is mediated by a tripartite ATP-independent periplasmic transporter.J Bacteriol. 2006 Dec;188(24):8407-12. doi: 10.1128/JB.00880-06. Epub 2006 Oct 13. J Bacteriol. 2006. PMID: 17041053 Free PMC article.
-
Dehalogenation of 4-chlorobenzoate. Characterisation of 4-chlorobenzoyl-coenzyme A dehalogenase from Pseudomonas sp. CBS3.Biodegradation. 1995 Sep;6(3):203-12. doi: 10.1007/BF00700458. Biodegradation. 1995. PMID: 7579994
-
Reductive, coenzyme A-mediated pathway for 3-chlorobenzoate degradation in the phototrophic bacterium Rhodopseudomonas palustris.Appl Environ Microbiol. 2001 Mar;67(3):1396-9. doi: 10.1128/AEM.67.3.1396-1399.2001. Appl Environ Microbiol. 2001. PMID: 11229940 Free PMC article.
-
Cloning and sequence analysis of genes for dehalogenation of 4-chlorobenzoate from Arthrobacter sp. strain SU.Appl Environ Microbiol. 1992 Dec;58(12):4068-71. doi: 10.1128/aem.58.12.4068-4071.1992. Appl Environ Microbiol. 1992. PMID: 1476446 Free PMC article.
Cited by
-
DipR, a GntR/FadR-family transcriptional repressor: regulatory mechanism and widespread distribution of the dip cluster for dipicolinic acid catabolism in bacteria.Nucleic Acids Res. 2024 Oct 14;52(18):10951-10964. doi: 10.1093/nar/gkae728. Nucleic Acids Res. 2024. PMID: 39180394 Free PMC article.
-
Regulation Mechanism of Nicotine Catabolism in Sphingomonas melonis TY by a Dual Role Transcriptional Regulator NdpR.Appl Environ Microbiol. 2023 May 31;89(5):e0032423. doi: 10.1128/aem.00324-23. Epub 2023 Apr 18. Appl Environ Microbiol. 2023. PMID: 37071026 Free PMC article.
-
PcaR, a GntR/FadR Family Transcriptional Repressor Controls the Transcription of Phenazine-1-Carboxylic Acid 1,2-Dioxygenase Gene Cluster in Sphingomonas histidinilytica DS-9.Appl Environ Microbiol. 2023 Jun 28;89(6):e0212122. doi: 10.1128/aem.02121-22. Epub 2023 May 16. Appl Environ Microbiol. 2023. PMID: 37191535 Free PMC article.
-
The TetR Family Repressor HpaR Negatively Regulates the Catabolism of 5-Hydroxypicolinic Acid in Alcaligenes faecalis JQ135 by Binding to Two Unique DNA Sequences in the Promoter of Hpa Operon.Appl Environ Microbiol. 2022 Mar 22;88(6):e0239021. doi: 10.1128/aem.02390-21. Epub 2022 Feb 9. Appl Environ Microbiol. 2022. PMID: 35138929 Free PMC article.
-
Unveiling the regulatory mechanisms of salicylate degradation gene cluster cehGHIR4 in Rhizobium sp. strain X9.Appl Environ Microbiol. 2023 Oct 31;89(10):e0080223. doi: 10.1128/aem.00802-23. Epub 2023 Oct 6. Appl Environ Microbiol. 2023. PMID: 37800922 Free PMC article.
References
-
- Chu CW, Liu B, Li N, Yao SG, Cheng D, Zhao JD, Qiu JG, Yan X, He Q, He J. 2017. A novel aerobic degradation pathway for thiobencarb is initiated by the TmoAB two-component flavin mononucleotide-dependent monooxygenase system in Acidovorax sp. strain T1. Appl Environ Microbiol 83:e01490-17. doi:10.1128/AEM.01490-17. - DOI - PMC - PubMed
-
- Baggi G. 2002. Microbial degradation of chlorobenzoates (CBAs): biochemical aspects and ecological implications, vol 36. Elsevier, Amsterdam, Netherlands.
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources

