Harnessing CRISPR-Cas9 for Genome Editing in Streptococcus pneumoniae D39V
- PMID: 33397704
- PMCID: PMC8105017
- DOI: 10.1128/AEM.02762-20
Harnessing CRISPR-Cas9 for Genome Editing in Streptococcus pneumoniae D39V
Abstract
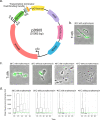
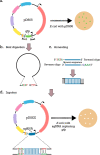
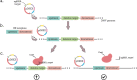
CRISPR-Cas systems provide bacteria and archaea with adaptive immunity against viruses and plasmids by the detection and cleavage of invading foreign DNA. Modified versions of this system can be exploited as a biotechnological tool for precise genome editing at a targeted locus. Here, we developed a replicative plasmid that carries the CRISPR-Cas9 system for RNA-programmable genome editing by counterselection in the opportunistic human pathogen Streptococcus pneumoniae Specifically, we demonstrate an approach for making targeted markerless gene knockouts and large genome deletions. After a precise double-stranded break (DSB) is introduced, the cells' DNA repair mechanism of homology-directed repair (HDR) is exploited to select successful transformants. This is achieved through the transformation of a template DNA fragment that will recombine in the genome and eliminate recognition of the target of the Cas9 endonuclease. Next, the newly engineered strain can be easily cured from the plasmid, which is temperature sensitive for replication, by growing it at the nonpermissive temperature. This allows for consecutive rounds of genome editing. Using this system, we engineered a strain with three major virulence factors deleted. The approaches developed here could potentially be adapted for use with other Gram-positive bacteria.IMPORTANCEStreptococcus pneumoniae (the pneumococcus) is an important opportunistic human pathogen killing more than 1 million people each year. Having the availability of a system capable of easy genome editing would significantly facilitate drug discovery and efforts to identify new vaccine candidates. Here, we introduced an easy-to-use system to perform multiple rounds of genome editing in the pneumococcus by putting the CRISPR-Cas9 system on a temperature-sensitive replicative plasmid. The approaches used here will advance genome editing projects in this important human pathogen.
Keywords: CRISPR; Cas9; Streptococcus pneumoniae; genome editing; plasmids.
Copyright © 2021 Synefiaridou and Veening.
Figures


Similar articles
-
Make-or-break prime editing for genome engineering in Streptococcus pneumoniae.Nat Commun. 2025 Apr 23;16(1):3796. doi: 10.1038/s41467-025-59068-8. Nat Commun. 2025. PMID: 40263274 Free PMC article.
-
CRISPR-Cas9 and CRISPR-Assisted Cytidine Deaminase Enable Precise and Efficient Genome Editing in Klebsiella pneumoniae.Appl Environ Microbiol. 2018 Nov 15;84(23):e01834-18. doi: 10.1128/AEM.01834-18. Print 2018 Dec 1. Appl Environ Microbiol. 2018. PMID: 30217854 Free PMC article.
-
Editing of the Bacillus subtilis Genome by the CRISPR-Cas9 System.Appl Environ Microbiol. 2016 Aug 15;82(17):5421-7. doi: 10.1128/AEM.01453-16. Print 2016 Sep 1. Appl Environ Microbiol. 2016. PMID: 27342565 Free PMC article.
-
Precision genome editing in the CRISPR era.Biochem Cell Biol. 2017 Apr;95(2):187-201. doi: 10.1139/bcb-2016-0137. Epub 2016 Sep 29. Biochem Cell Biol. 2017. PMID: 28177771 Review.
-
Optimization of genome editing through CRISPR-Cas9 engineering.Bioengineered. 2016 Apr;7(3):166-74. doi: 10.1080/21655979.2016.1189039. Bioengineered. 2016. PMID: 27340770 Free PMC article. Review.
Cited by
-
Amoxicillin-resistant Streptococcus pneumoniae can be resensitized by targeting the mevalonate pathway as indicated by sCRilecs-seq.Elife. 2022 Jun 24;11:e75607. doi: 10.7554/eLife.75607. Elife. 2022. PMID: 35748540 Free PMC article.
-
Inhibition of pneumococcal growth and biofilm formation by human isolates of Streptococcus mitis and Streptococcus oralis.Appl Environ Microbiol. 2025 Mar 19;91(3):e0133624. doi: 10.1128/aem.01336-24. Epub 2025 Feb 26. Appl Environ Microbiol. 2025. PMID: 40008876 Free PMC article.
-
Characterization of NiCas12b for In Vivo Genome Editing.Adv Sci (Weinh). 2024 Sep;11(36):e2400469. doi: 10.1002/advs.202400469. Epub 2024 Jul 30. Adv Sci (Weinh). 2024. PMID: 39076074 Free PMC article.
-
Group B Streptococcus Cas9 variants provide insight into programmable gene repression and CRISPR-Cas transcriptional effects.Commun Biol. 2023 Jun 9;6(1):620. doi: 10.1038/s42003-023-04994-w. Commun Biol. 2023. PMID: 37296208 Free PMC article.
-
Cotton under heat stress: a comprehensive review of molecular breeding, genomics, and multi-omics strategies.Front Genet. 2025 Mar 18;16:1553406. doi: 10.3389/fgene.2025.1553406. eCollection 2025. Front Genet. 2025. PMID: 40171219 Free PMC article. Review.
References
-
- O'Brien KL, Wolfson LJ, Watt JP, Henkle E, Deloria-Knoll M, McCall N, Lee E, Mulholland K, Levine OS, Cherian T, Hib and Pneumococcal Global Burden of Disease Study Team. 2009. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374:893–902. doi:10.1016/S0140-6736(09)61204-6. - DOI - PubMed
-
- Avery OT, MacLeod CM, McCarty M. 1944. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J Exp Med 79:137–158. doi:10.1084/jem.79.2.137. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

