A RAC-GEF network critical for early intestinal tumourigenesis
- PMID: 33397922
- PMCID: PMC7782582
- DOI: 10.1038/s41467-020-20255-4
A RAC-GEF network critical for early intestinal tumourigenesis
Abstract
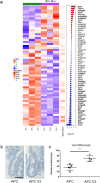
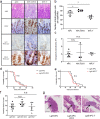
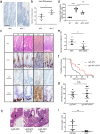
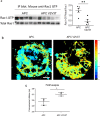
RAC1 activity is critical for intestinal homeostasis, and is required for hyperproliferation driven by loss of the tumour suppressor gene Apc in the murine intestine. To avoid the impact of direct targeting upon homeostasis, we reasoned that indirect targeting of RAC1 via RAC-GEFs might be effective. Transcriptional profiling of Apc deficient intestinal tissue identified Vav3 and Tiam1 as key targets. Deletion of these indicated that while TIAM1 deficiency could suppress Apc-driven hyperproliferation, it had no impact upon tumourigenesis, while VAV3 deficiency had no effect. Intriguingly, deletion of either gene resulted in upregulation of Vav2, with subsequent targeting of all three (Vav2-/- Vav3-/- Tiam1-/-), profoundly suppressing hyperproliferation, tumourigenesis and RAC1 activity, without impacting normal homeostasis. Critically, the observed RAC-GEF dependency was negated by oncogenic KRAS mutation. Together, these data demonstrate that while targeting RAC-GEF molecules may have therapeutic impact at early stages, this benefit may be lost in late stage disease.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
- G117/424/MRC_/Medical Research Council/United Kingdom
- 20410/CRUK_/Cancer Research UK/United Kingdom
- A15673/CRUK_/Cancer Research UK/United Kingdom
- A29055/CRUK_/Cancer Research UK/United Kingdom
- BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- A17196/CRUK_/Cancer Research UK/United Kingdom
- 21139/CRUK_/Cancer Research UK/United Kingdom
- 24452/CRUK_/Cancer Research UK/United Kingdom
- 19166/CRUK_/Cancer Research UK/United Kingdom
- A25045/CRUK_/Cancer Research UK/United Kingdom
- A15565/CRUK_/Cancer Research UK/United Kingdom
- A21139/CRUK_/Cancer Research UK/United Kingdom
- 25808/CRUK_/Cancer Research UK/United Kingdom
- A17096/CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

