The order and logic of CD4 versus CD8 lineage choice and differentiation in mouse thymus
- PMID: 33397934
- PMCID: PMC7782583
- DOI: 10.1038/s41467-020-20306-w
The order and logic of CD4 versus CD8 lineage choice and differentiation in mouse thymus
Abstract
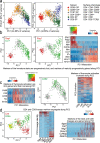
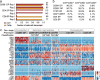
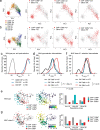
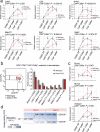
CD4 and CD8 mark helper and cytotoxic T cell lineages, respectively, and serve as coreceptors for MHC-restricted TCR recognition. How coreceptor expression is matched with TCR specificity is central to understanding CD4/CD8 lineage choice, but visualising coreceptor gene activity in individual selection intermediates has been technically challenging. It therefore remains unclear whether the sequence of coreceptor gene expression in selection intermediates follows a stereotypic pattern, or is responsive to signaling. Here we use single cell RNA sequencing (scRNA-seq) to classify mouse thymocyte selection intermediates by coreceptor gene expression. In the unperturbed thymus, Cd4+Cd8a- selection intermediates appear before Cd4-Cd8a+ selection intermediates, but the timing of these subsets is flexible according to the strength of TCR signals. Our data show that selection intermediates discriminate MHC class prior to the loss of coreceptor expression and suggest a model where signal strength informs the timing of coreceptor gene activity and ultimately CD4/CD8 lineage choice.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

