Site-specific incorporation of citrulline into proteins in mammalian cells
- PMID: 33398026
- PMCID: PMC7782748
- DOI: 10.1038/s41467-020-20279-w
Site-specific incorporation of citrulline into proteins in mammalian cells
Abstract
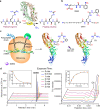
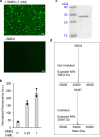
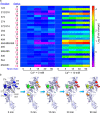
Citrullination is a post-translational modification (PTM) of arginine that is crucial for several physiological processes, including gene regulation and neutrophil extracellular trap formation. Despite recent advances, studies of protein citrullination remain challenging due to the difficulty of accessing proteins homogeneously citrullinated at a specific site. Herein, we report a technology that enables the site-specific incorporation of citrulline (Cit) into proteins in mammalian cells. This approach exploits an engineered E. coli-derived leucyl tRNA synthetase-tRNA pair that incorporates a photocaged-citrulline (SM60) into proteins in response to a nonsense codon. Subsequently, SM60 is readily converted to Cit with light in vitro and in living cells. To demonstrate the utility of the method, we biochemically characterize the effect of incorporating Cit at two known autocitrullination sites in Protein Arginine Deiminase 4 (PAD4, R372 and R374) and show that the R372Cit and R374Cit mutants are 181- and 9-fold less active than the wild-type enzyme. This technology possesses the potential to decipher the biology of citrullination.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

