DNA-protein crosslink proteases in genome stability
- PMID: 33398053
- PMCID: PMC7782752
- DOI: 10.1038/s42003-020-01539-3
DNA-protein crosslink proteases in genome stability
Abstract
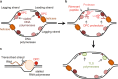
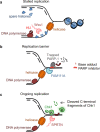
Proteins covalently attached to DNA, also known as DNA-protein crosslinks (DPCs), are common and bulky DNA lesions that interfere with DNA replication, repair, transcription and recombination. Research in the past several years indicates that cells possess dedicated enzymes, known as DPC proteases, which digest the protein component of a DPC. Interestingly, DPC proteases also play a role in proteolysis beside DPC repair, such as in degrading excess histones during DNA replication or controlling DNA replication checkpoints. Here, we discuss the importance of DPC proteases in DNA replication, genome stability and their direct link to human diseases and cancer therapy.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

