Homer1 promotes dendritic spine growth through ankyrin-G and its loss reshapes the synaptic proteome
- PMID: 33398084
- PMCID: PMC8254828
- DOI: 10.1038/s41380-020-00991-1
Homer1 promotes dendritic spine growth through ankyrin-G and its loss reshapes the synaptic proteome
Abstract
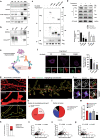
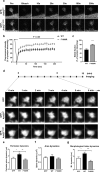
Homer1 is a synaptic scaffold protein that regulates glutamatergic synapses and spine morphogenesis. HOMER1 knockout (KO) mice show behavioral abnormalities related to psychiatric disorders, and HOMER1 has been associated with psychiatric disorders such as addiction, autism disorder (ASD), schizophrenia (SZ), and depression. However, the mechanisms by which it promotes spine stability and its global function in maintaining the synaptic proteome has not yet been fully investigated. Here, we used computational approaches to identify global functions for proteins containing the Homer1-interacting PPXXF motif within the postsynaptic compartment. Ankyrin-G was one of the most topologically important nodes in the postsynaptic peripheral membrane subnetwork, and we show that one of the PPXXF motifs, present in the postsynaptically-enriched 190 kDa isoform of ankyrin-G (ankyrin-G 190), is recognized by the EVH1 domain of Homer1. We use proximity ligation combined with super-resolution microscopy to map the interaction of ankyrin-G and Homer1 to distinct nanodomains within the spine head and correlate them with spine head size. This interaction motif is critical for ankyrin-G 190's ability to increase spine head size, and for the maintenance of a stable ankyrin-G pool in spines. Intriguingly, lack of Homer1 significantly upregulated the abundance of ankyrin-G, but downregulated Shank3 in cortical crude plasma membrane fractions. In addition, proteomic analysis of the cortex in HOMER1 KO and wild-type (WT) mice revealed a global reshaping of the postsynaptic proteome, surprisingly characterized by extensive upregulation of synaptic proteins. Taken together, we show that Homer1 and its protein interaction motif have broad global functions within synaptic protein-protein interaction networks. Enrichment of disease risk factors within these networks has important implications for neurodevelopmental disorders including bipolar disorder, ASD, and SZ.
© 2021. The Author(s), under exclusive licence to Springer Nature Limited part of Springer Nature.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

