Systemic viral spreading and defective host responses are associated with fatal Lassa fever in macaques
- PMID: 33398113
- PMCID: PMC7782745
- DOI: 10.1038/s42003-020-01543-7
Systemic viral spreading and defective host responses are associated with fatal Lassa fever in macaques
Abstract
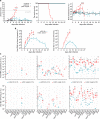
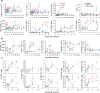
Lassa virus (LASV) is endemic in West Africa and induces a viral hemorrhagic fever (VHF) with up to 30% lethality among clinical cases. The mechanisms involved in control of Lassa fever or, in contrast, the ensuing catastrophic illness and death are poorly understood. We used the cynomolgus monkey model to reproduce the human disease with asymptomatic to mild or fatal disease. After initial replication at the inoculation site, LASV reached the secondary lymphoid organs. LASV did not spread further in nonfatal disease and was rapidly controlled by balanced innate and T-cell responses. Systemic viral dissemination occurred during severe disease. Massive replication, a cytokine/chemokine storm, defective T-cell responses, and multiorgan failure were observed. Clinical, biological, immunological, and transcriptomic parameters resembled those observed during septic-shock syndrome, suggesting that similar pathogenesis is induced during Lassa fever. The outcome appears to be determined early, as differentially expressed genes in PBMCs were associated with fatal and non-fatal Lassa fever outcome very early after infection. These results provide a full characterization and important insights into Lassa fever pathogenesis and could help to develop early diagnostic tools.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures








References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

