Single-cell dissection of intratumoral heterogeneity and lineage diversity in metastatic gastric adenocarcinoma
- PMID: 33398161
- PMCID: PMC8074162
- DOI: 10.1038/s41591-020-1125-8
Single-cell dissection of intratumoral heterogeneity and lineage diversity in metastatic gastric adenocarcinoma
Abstract
Intratumoral heterogeneity (ITH) is a fundamental property of cancer; however, the origins of ITH remain poorly understood. We performed single-cell transcriptome profiling of peritoneal carcinomatosis (PC) from 15 patients with gastric adenocarcinoma (GAC), constructed a map of 45,048 PC cells, profiled the transcriptome states of tumor cell populations, incisively explored ITH of malignant PC cells and identified significant correlates with patient survival. The links between tumor cell lineage/state compositions and ITH were illustrated at transcriptomic, genotypic, molecular and phenotypic levels. We uncovered the diversity in tumor cell lineage/state compositions in PC specimens and defined it as a key contributor to ITH. Single-cell analysis of ITH classified PC specimens into two subtypes that were prognostically independent of clinical variables, and a 12-gene prognostic signature was derived and validated in multiple large-scale GAC cohorts. The prognostic signature appears fundamental to GAC carcinogenesis and progression and could be practical for patient stratification.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures

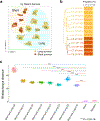
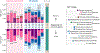
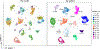











References
-
- Bray F et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin 68, 394–424 (2018). - PubMed
-
- Shiozaki H et al. Prognosis of gastric adenocarcinoma patients with various burdens of peritoneal metastases. J. Surg. Oncol 113, 29–35 (2016). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

