NF-κB-inducing kinase maintains T cell metabolic fitness in antitumor immunity
- PMID: 33398181
- PMCID: PMC7855506
- DOI: 10.1038/s41590-020-00829-6
NF-κB-inducing kinase maintains T cell metabolic fitness in antitumor immunity
Erratum in
-
Author Correction: NF-κB-inducing kinase maintains T cell metabolic fitness in antitumor immunity.Nat Immunol. 2021 Apr;22(4):530. doi: 10.1038/s41590-021-00892-7. Nat Immunol. 2021. PMID: 33574621 No abstract available.
Abstract
Metabolic reprograming toward aerobic glycolysis is a pivotal mechanism shaping immune responses. Here we show that deficiency in NF-κB-inducing kinase (NIK) impairs glycolysis induction, rendering CD8+ effector T cells hypofunctional in the tumor microenvironment. Conversely, ectopic expression of NIK promotes CD8+ T cell metabolism and effector function, thereby profoundly enhancing antitumor immunity and improving the efficacy of T cell adoptive therapy. NIK regulates T cell metabolism via a NF-κB-independent mechanism that involves stabilization of hexokinase 2 (HK2), a rate-limiting enzyme of the glycolytic pathway. NIK prevents autophagic degradation of HK2 through controlling cellular reactive oxygen species levels, which in turn involves modulation of glucose-6-phosphate dehydrogenase (G6PD), an enzyme that mediates production of the antioxidant NADPH. We show that the G6PD-NADPH redox system is important for HK2 stability and metabolism in activated T cells. These findings establish NIK as a pivotal regulator of T cell metabolism and highlight a post-translational mechanism of metabolic regulation.
Figures

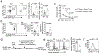
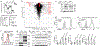

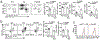
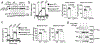


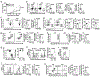




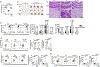
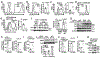
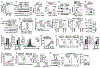
References
-
- Almeida L, Lochner M, Berod L & Sparwasser T Metabolic pathways in T cell activation and lineage differentiation. Semin Immunol 28, 514–524 (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

