This is a preprint.
Structural basis for broad coronavirus neutralization
- PMID: 33398277
- PMCID: PMC7781312
- DOI: 10.1101/2020.12.29.424482
Structural basis for broad coronavirus neutralization
Update in
-
Structural basis for broad coronavirus neutralization.Nat Struct Mol Biol. 2021 Jun;28(6):478-486. doi: 10.1038/s41594-021-00596-4. Epub 2021 May 12. Nat Struct Mol Biol. 2021. PMID: 33981021
Abstract
Three highly pathogenic β-coronaviruses crossed the animal-to-human species barrier in the past two decades: SARS-CoV, MERS-CoV and SARS-CoV-2. SARS-CoV-2 has infected more than 64 million people worldwide, claimed over 1.4 million lives and is responsible for the ongoing COVID-19 pandemic. We isolated a monoclonal antibody, termed B6, cross-reacting with eight β-coronavirus spike glycoproteins, including all five human-infecting β-coronaviruses, and broadly inhibiting entry of pseudotyped viruses from two coronavirus lineages. Cryo-electron microscopy and X-ray crystallography characterization reveal that B6 binds to a conserved cryptic epitope located in the fusion machinery and indicate that antibody binding sterically interferes with spike conformational changes leading to membrane fusion. Our data provide a structural framework explaining B6 cross-reactivity with β-coronaviruses from three lineages along with proof-of-concept for antibody-mediated broad coronavirus neutralization elicited through vaccination. This study unveils an unexpected target for next-generation structure-guided design of a pan-coronavirus vaccine.
Conflict of interest statement
Declaration of interests M.M.S, M.A.T., Y.J.P., A.C.W, A.T.M. and D.V. are named as inventors on patent applications filed by the University of Washington based on the studies presented in this paper. D.V. is a consultant for Vir Biotechnology Inc. The Veesler laboratory has received an unrelated sponsored research agreement from Vir Biotechnology Inc. The other authors declare no competing interests.
Figures

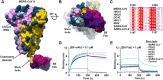


References
-
- Agirre J., Iglesias-Fernández J., Rovira C., Davies G.J., Wilson K.S., and Cowtan K.D. (2015). Privateer: software for the conformational validation of carbohydrate structures. Nat Struct Mol Biol 22, 833–834. - PubMed
-
- Azoitei M.L., Correia B.E., Ban Y.E., Carrico C., Kalyuzhniy O., Chen L., Schroeter A., Huang P.S., McLellan J.S., Kwong P.D., et al. (2011). Computation-guided backbone grafting of a discontinuous motif onto a protein scaffold. Science 334, 373–376. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
