This is a preprint.
REAL-TIME MECHANISTIC BAYESIAN FORECASTS OF COVID-19 MORTALITY
- PMID: 33398305
- PMCID: PMC7781348
- DOI: 10.1101/2020.12.22.20248736
REAL-TIME MECHANISTIC BAYESIAN FORECASTS OF COVID-19 MORTALITY
Update in
-
REAL-TIME MECHANISTIC BAYESIAN FORECASTS OF COVID-19 MORTALITY.Ann Appl Stat. 2023 Sep;17(3):1801-1819. doi: 10.1214/22-aoas1671. Epub 2023 Sep 7. Ann Appl Stat. 2023. PMID: 38983109 Free PMC article.
Abstract
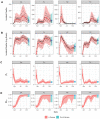
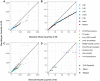
The COVID-19 pandemic emerged in late December 2019. In the first six months of the global outbreak, the US reported more cases and deaths than any other country in the world. Effective modeling of the course of the pandemic can help assist with public health resource planning, intervention efforts, and vaccine clinical trials. However, building applied forecasting models presents unique challenges during a pandemic. First, case data available to models in real-time represent a non-stationary fraction of the true case incidence due to changes in available diagnostic tests and test-seeking behavior. Second, interventions varied across time and geography leading to large changes in transmissibility over the course of the pandemic. We propose a mechanistic Bayesian model (MechBayes) that builds upon the classic compartmental susceptible-exposed-infected-recovered (SEIR) model to operationalize COVID-19 forecasting in real time. This framework includes non-parametric modeling of varying transmission rates, non-parametric modeling of case and death discrepancies due to testing and reporting issues, and a joint observation likelihood on new case counts and new deaths; it is implemented in a probabilistic programming language to automate the use of Bayesian reasoning for quantifying uncertainty in probabilistic forecasts. The model has been used to submit forecasts to the US Centers for Disease Control, through the COVID-19 Forecast Hub. We examine the performance relative to a baseline model as well as alternate models submitted to the Forecast Hub. Additionally, we include an ablation test of our extensions to the classic SEIR model. We demonstrate a significant gain in both point and probabilistic forecast scoring measures using MechBayes when compared to a baseline model and show that MechBayes ranks as one of the top 2 models out of 10 submitted to the COVID-19 Forecast Hub. Finally, we demonstrate that MechBayes performs significantly better than the classical SEIR model.
Figures



References
-
- Ray Evan L, Wattanachit Nutcha, Niemi Jarad, Kanji Abdul Hannan, House Katie, Cramer Estee Y, Bracher Johannes, Zheng Andrew, Yamana Teresa K, Xiong Xinyue, et al. Ensemble Forecasts of Coronavirus Disease 2019 (COVID-19) in the us. medRxiv, 2020.
-
- Lutz Chelsea S, Huynh Mimi P, Schroeder Monica, Anyatonwu Sophia, Dahlgren F Scott, Danyluk Gregory, Fernandez Danielle, Greene Sharon K, Kipshidze Nodar, Liu Leann, et al. Applying infectious disease forecasting to public health: a path forward using influenza forecasting examples. BMC Public Health, 19(1):1659, 2019. - PMC - PubMed
-
- Kermack William Ogilvy and McKendrick Anderson G. A contribution to the mathematical theory of epidemics. Proceedings of the royal society of london. Series A, Containing papers of a mathematical and physical character, 115(772):700–721, 1927.
-
- Korolev Ivan. Identification and Estimation of the SEIRD epidemic model for COVID-19. Binghamton University. http://dx.doi.org/10.2139/ssrn, 3569367, 2020. - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
