Identification of KIF4A and its effect on the progression of lung adenocarcinoma based on the bioinformatics analysis
- PMID: 33398330
- PMCID: PMC7823194
- DOI: 10.1042/BSR20203973
Identification of KIF4A and its effect on the progression of lung adenocarcinoma based on the bioinformatics analysis
Abstract
Background: Lung adenocarcinoma (LUAD) is the most frequent histological type of lung cancer, and its incidence has displayed an upward trend in recent years. Nevertheless, little is known regarding effective biomarkers for LUAD.
Methods: The robust rank aggregation method was used to mine differentially expressed genes (DEGs) from the gene expression omnibus (GEO) datasets. The Search Tool for the Retrieval of Interacting Genes (STRING) database was used to extract hub genes from the protein-protein interaction (PPI) network. The expression of the hub genes was validated using expression profiles from TCGA and Oncomine databases and was verified by real-time quantitative PCR (qRT-PCR). The module and survival analyses of the hub genes were determined using Cytoscape and Kaplan-Meier curves. The function of KIF4A as a hub gene was investigated in LUAD cell lines.
Results: The PPI analysis identified seven DEGs including BIRC5, DLGAP5, CENPF, KIF4A, TOP2A, AURKA, and CCNA2, which were significantly upregulated in Oncomine and TCGA LUAD datasets, and were verified by qRT-PCR in our clinical samples. We determined the overall and disease-free survival analysis of the seven hub genes using GEPIA. We further found that CENPF, DLGAP5, and KIF4A expressions were positively correlated with clinical stage. In LUAD cell lines, proliferation and migration were inhibited and apoptosis was promoted by knocking down KIF4A expression.
Conclusion: We have identified new DEGs and functional pathways involved in LUAD. KIF4A, as a hub gene, promoted the progression of LUAD and might represent a potential therapeutic target for molecular cancer therapy.
Keywords: Bioinformatical analysis; Biomarker; Differentially expressed genes; KIF4A; Lung adenocarcinoma.
© 2021 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures


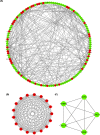


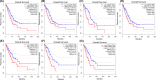




References
-
- Gazdar A.F. and Zhou C. (2018) Lung cancer in never-smokers: a different disease. IASLC Thoracic Oncology, pp. 23.e3–29.e3, Elsevier
-
- Liu Z., Liu B. and Li C. (2019) A case report of metastatic bilateral ovarian cancer due to non-small cell lung cancer with ALK gene rearrangement. Eur. J. Gynaecol. Oncol. 40, 151–153
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

