A combinatorial method to visualize the neuronal network in the mouse spinal cord: combination of a modified Golgi-Cox method and synchrotron radiation micro-computed tomography
- PMID: 33398435
- PMCID: PMC8062354
- DOI: 10.1007/s00418-020-01949-8
A combinatorial method to visualize the neuronal network in the mouse spinal cord: combination of a modified Golgi-Cox method and synchrotron radiation micro-computed tomography
Abstract
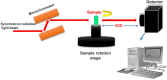
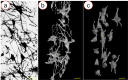
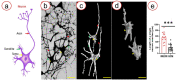
Exploring the three-dimensional (3D) morphology of neurons is essential to understanding spinal cord function and associated diseases comprehensively. However, 3D imaging of the neuronal network in the broad region of the spinal cord at cellular resolution remains a challenge in the field of neuroscience. In this study, to obtain high-resolution 3D imaging of a detailed neuronal network in the mass of the spinal cord, the combination of synchrotron radiation micro-computed tomography (SRμCT) and the Golgi-cox staining were used. We optimized the Golgi-Cox method (GCM) and developed a modified GCM (M-GCM), which improved background staining, reduced the number of artefacts, and diminished the impact of incomplete vasculature compared to the current GCM. Moreover, we achieved high-resolution 3D imaging of the detailed neuronal network in the spinal cord through the combination of SRμCT and M-GCM. Our results showed that the M-GCM increased the contrast between the neuronal structure and its surrounding extracellular matrix. Compared to the GCM, the M-GCM also diminished the impact of the artefacts and incomplete vasculature on the 3D image. Additionally, the 3D neuronal architecture was successfully quantified using a combination of SRμCT and M-GCM. The SRμCT was shown to be a valuable non-destructive tool for 3D visualization of the neuronal network in the broad 3D region of the spinal cord. Such a combinatorial method will, therefore, transform the presentation of Golgi staining from 2 to 3D, providing significant improvements in the 3D rendering of the neuronal network.
Keywords: Modified Golgi-Cox method; Neuronal network; SRμCT; Spinal cord; Three-dimension.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Cedola A, Bravin A, Bukreeva I, Fratini M, Pacureanu A, Mittone A, Massimi L, Cloetens P, Coan P, Campi G, Spano R, Brun F, Grigoryev V, Petrosino V, Venturi C, Mastrogiacomo M, Kerlero de Rosbo N, Uccelli A. X-ray phase contrast tomography reveals early vascular alterations and neuronal loss in a multiple sclerosis model. Sci Rep. 2017;7(1):5890. doi: 10.1038/s41598-017-06251-7. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

