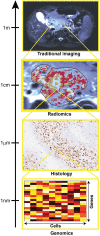
The Biological Meaning of Radiomic Features
- PMID: 33399513
- PMCID: PMC7924519
- DOI: 10.1148/radiol.2021202553
The Biological Meaning of Radiomic Features
Erratum in
-
The Biological Meaning of Radiomic Features.Radiology. 2021 May;299(2):E256. doi: 10.1148/radiol.2021219005. Radiology. 2021. PMID: 33900879 Free PMC article. No abstract available.
Abstract
An earlier incorrect version appeared online. This article was corrected on February 10, 2021.
Figures




References
-
- Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 2017;14(12):749–762. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

