Nucleic acid sample preparation from whole blood in a paper microfluidic device using isotachophoresis
- PMID: 33401049
- PMCID: PMC8115986
- DOI: 10.1016/j.jchromb.2020.122494
Nucleic acid sample preparation from whole blood in a paper microfluidic device using isotachophoresis
Abstract
Nucleic acid amplification tests (NAATs) are a crucial diagnostic and monitoring tool for infectious diseases. A key procedural step for NAATs is sample preparation: separating and purifying target nucleic acids from crude biological samples prior to nucleic acid amplification and detection. Traditionally, sample preparation has been performed with liquid- or solid-phase extraction, both of which require multiple trained user steps and significant laboratory equipment. The challenges associated with sample preparation have limited the dissemination of NAAT point-of-care diagnostics in low resource environments, including low- and middle-income countries. We report on a paper-based device for purification of nucleic acids from whole blood using isotachophoresis (ITP) for point-of-care NAATs. We show successful extraction and purification of target nucleic acids from large volumes (33 µL) of whole human blood samples with no moving parts and few user steps. Our device utilizes paper-based buffer reservoirs to fully contain the liquid ITP buffers and does not require complex filling procedures, instead relying on the natural wicking of integrated paper membranes. We perform on-device blood fractionation via filtration to remove leukocytes and erythrocytes from our sample, followed by integrated on-paper proteolytic digestion of endogenous plasma proteins to allow for successful isotachophoretic extraction. Paper-based isotachophoresis purifies and concentrates target nucleic acids that are added directly to recombinase polymerase amplification (RPA) reactions. We show consistent amplification of input copy concentrations of as low as 3 × 103 copies nucleic acid per mL input blood with extraction and purification taking only 30 min. By employing a paper architecture, we are able to incorporate these processes in a single, robust, low-cost design, enabling the direct processing of large volumes of blood, with the only intermediate user steps being the removal and addition of tape. Our device represents a step towards a simple, fully integrated sample preparation system for nucleic acid amplification tests at the point-of-care.
Keywords: Isotachophoresis; Nucleic acids; Paper-microfluidics; Protein digestion; Sample preparation.
Copyright © 2020 Elsevier B.V. All rights reserved.
Figures

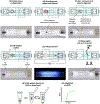

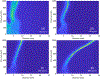
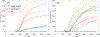
Similar articles
-
Semiquantitative Nucleic Acid Test with Simultaneous Isotachophoretic Extraction and Amplification.Anal Chem. 2018 Jun 19;90(12):7221-7229. doi: 10.1021/acs.analchem.8b00185. Epub 2018 May 25. Anal Chem. 2018. PMID: 29761701 Free PMC article.
-
NAIL: Nucleic Acid detection using Isotachophoresis and Loop-mediated isothermal amplification.Lab Chip. 2015 Apr 7;15(7):1697-707. doi: 10.1039/c4lc01479k. Lab Chip. 2015. PMID: 25666345
-
An injection molded microchip for nucleic acid purification from 25 microliter samples using isotachophoresis.J Chromatogr A. 2014 Feb 28;1331:139-42. doi: 10.1016/j.chroma.2014.01.036. Epub 2014 Jan 21. J Chromatogr A. 2014. PMID: 24485540
-
Recent progress in nucleic acids isotachophoresis.J Sep Sci. 2018 Jan;41(1):236-247. doi: 10.1002/jssc.201700878. Epub 2017 Oct 30. J Sep Sci. 2018. PMID: 28980403 Review.
-
Paper microfluidics for nucleic acid amplification testing (NAAT) of infectious diseases.Lab Chip. 2017 Jul 11;17(14):2347-2371. doi: 10.1039/c7lc00013h. Lab Chip. 2017. PMID: 28632278 Review.
Cited by
-
Recent Advances in Microfluidic Devices for Contamination Detection and Quality Inspection of Milk.Micromachines (Basel). 2021 May 14;12(5):558. doi: 10.3390/mi12050558. Micromachines (Basel). 2021. PMID: 34068982 Free PMC article. Review.
-
Simple Electric Device to Isolate Nucleic Acids from Whole Blood Optimized for Point of Care Testing of Brain Damage.Curr Neurovasc Res. 2022;19(3):333-343. doi: 10.2174/1567202619666220903105805. Curr Neurovasc Res. 2022. PMID: 36056832 Free PMC article.
-
CRISPR Assays for Disease Diagnosis: Progress to and Barriers Remaining for Clinical Applications.Adv Sci (Weinh). 2023 Jul;10(20):e2301697. doi: 10.1002/advs.202301697. Epub 2023 May 10. Adv Sci (Weinh). 2023. PMID: 37162202 Free PMC article. Review.
-
Research on a Magnetic Separation-Based Rapid Nucleic Acid Extraction System and Its Detection Applications.Biosensors (Basel). 2023 Sep 23;13(10):903. doi: 10.3390/bios13100903. Biosensors (Basel). 2023. PMID: 37887096 Free PMC article.
-
Self-Diagnosis of SARS-CoV-2 from Saliva Samples at Home: Isothermal Amplification Enabled by Do-It-Yourself Portable Incubators and Laminated Poly-ethyl Sulfonate Membranes.Diagnostics (Basel). 2024 Jan 19;14(2):221. doi: 10.3390/diagnostics14020221. Diagnostics (Basel). 2024. PMID: 38275468 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

