First Report of CRISPR/Cas9 Mediated DNA-Free Editing of 4CL and RVE7 Genes in Chickpea Protoplasts
- PMID: 33401455
- PMCID: PMC7795094
- DOI: 10.3390/ijms22010396
First Report of CRISPR/Cas9 Mediated DNA-Free Editing of 4CL and RVE7 Genes in Chickpea Protoplasts
Abstract
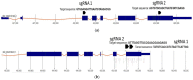
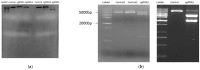
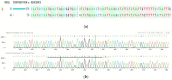
The current genome editing system Clustered Regularly Interspaced Short Palindromic Repeats Cas9 (CRISPR/Cas9) has already confirmed its proficiency, adaptability, and simplicity in several plant-based applications. Together with the availability of a vast amount of genome data and transcriptome data, CRISPR/Cas9 presents a massive opportunity for plant breeders and researchers. The successful delivery of ribonucleoproteins (RNPs), which are composed of Cas9 enzyme and a synthetically designed single guide RNA (sgRNA) and are used in combination with various transformation methods or lately available novel nanoparticle-based delivery approaches, allows targeted mutagenesis in plants species. Even though this editing technique is limitless, it has still not been employed in many plant species to date. Chickpea is the second most crucial winter grain crop cultivated worldwide; there are currently no reports on CRISPR/Cas9 gene editing in chickpea. Here, we selected the 4-coumarate ligase (4CL) and Reveille 7 (RVE7) genes, both associated with drought tolerance for CRISPR/Cas9 editing in chickpea protoplast. The 4CL represents a key enzyme involved in phenylpropanoid metabolism in the lignin biosynthesis pathway. It regulates the accumulation of lignin under stress conditions in several plants. The RVE7 is a MYB transcription factor which is part of regulating circadian rhythm in plants. The knockout of these selected genes in the chickpea protoplast using DNA-free CRISPR/Cas9 editing represents a novel approach for achieving targeted mutagenesis in chickpea. Results showed high-efficiency editing was achieved for RVE7 gene in vivo compared to the 4CL gene. This study will help unravel the role of these genes under drought stress and understand the complex drought stress mechanism pathways. This is the first study in chickpea protoplast utilizing CRISPR/Cas9 DNA free gene editing of drought tolerance associated genes.
Keywords: 4-coumarate ligase (4CL); CRISPR/Cas9; DNA free gene editing; Reveille 7 (RVE7); chickpea; drought stress; protoplast.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



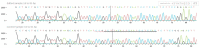
References
-
- Lin C.-S., Hsu C.-T., Yang L.-H., Lee L.-Y., Fu J.-Y., Cheng Q.-W., Wu F.-H., Hsiao H.C.-W., Zhang Y., Zhang R., et al. Application of protoplast technology to CRISPR/Cas9 mutagenesis: From single-cell mutation detection to mutant plant regeneration. Plant Biotechnol. J. 2018;16:1295–1310. doi: 10.1111/pbi.12870. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

