Master Regulators of Muscle Atrophy: Role of Costamere Components
- PMID: 33401549
- PMCID: PMC7823551
- DOI: 10.3390/cells10010061
Master Regulators of Muscle Atrophy: Role of Costamere Components
Abstract
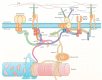
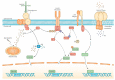
The loss of muscle mass and force characterizes muscle atrophy in several different conditions, which share the expression of atrogenes and the activation of their transcriptional regulators. However, attempts to antagonize muscle atrophy development in different experimental contexts by targeting contributors to the atrogene pathway showed partial effects in most cases. Other master regulators might independently contribute to muscle atrophy, as suggested by our recent evidence about the co-requirement of the muscle-specific chaperone protein melusin to inhibit unloading muscle atrophy development. Furthermore, melusin and other muscle mass regulators, such as nNOS, belong to costameres, the macromolecular complexes that connect sarcolemma to myofibrils and to the extracellular matrix, in correspondence with specific sarcomeric sites. Costameres sense a mechanical load and transduce it both as lateral force and biochemical signals. Recent evidence further broadens this classic view, by revealing the crucial participation of costameres in a sarcolemmal "signaling hub" integrating mechanical and humoral stimuli, where mechanical signals are coupled with insulin and/or insulin-like growth factor stimulation to regulate muscle mass. Therefore, this review aims to enucleate available evidence concerning the early involvement of costamere components and additional putative master regulators in the development of major types of muscle atrophy.
Keywords: aging; atrogene; cachexia; costamere; dystrophin; melusin; muscle atrophy; muscle disuse; nNOS; sarcopenia.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Loss of melusin is a novel, neuronal NO synthase/FoxO3-independent master switch of unloading-induced muscle atrophy.J Cachexia Sarcopenia Muscle. 2020 Jun;11(3):802-819. doi: 10.1002/jcsm.12546. Epub 2020 Mar 10. J Cachexia Sarcopenia Muscle. 2020. PMID: 32154658 Free PMC article.
-
Sarcolemmal loss of active nNOS (Nos1) is an oxidative stress-dependent, early event driving disuse atrophy.J Pathol. 2018 Dec;246(4):433-446. doi: 10.1002/path.5149. Epub 2018 Oct 24. J Pathol. 2018. PMID: 30066461
-
Evidence for skeletal muscle fiber type-specific expressions of mechanosensors.Cell Mol Life Sci. 2019 Aug;76(15):2987-3004. doi: 10.1007/s00018-019-03026-3. Epub 2019 Jan 30. Cell Mol Life Sci. 2019. PMID: 30701284 Free PMC article.
-
Chaperone Proteins: The Rising Players in Muscle Atrophy.J Cachexia Sarcopenia Muscle. 2025 Feb;16(1):e13659. doi: 10.1002/jcsm.13659. Epub 2024 Dec 20. J Cachexia Sarcopenia Muscle. 2025. PMID: 39707668 Free PMC article. Review.
-
Skeletal muscle atrophy: disease-induced mechanisms may mask disuse atrophy.J Muscle Res Cell Motil. 2015 Dec;36(6):405-21. doi: 10.1007/s10974-015-9439-8. Epub 2016 Jan 4. J Muscle Res Cell Motil. 2015. PMID: 26728748 Review.
Cited by
-
Delivery of engineered extracellular vesicles with miR-29b editing system for muscle atrophy therapy.J Nanobiotechnology. 2022 Jun 27;20(1):304. doi: 10.1186/s12951-022-01508-4. J Nanobiotechnology. 2022. PMID: 35761332 Free PMC article.
-
Blocking insulin-like growth factor 1 receptor signaling pathway inhibits neuromuscular junction regeneration after botulinum toxin-A treatment.Cell Death Dis. 2023 Sep 16;14(9):609. doi: 10.1038/s41419-023-06128-w. Cell Death Dis. 2023. PMID: 37717026 Free PMC article.
-
Phosphoproteomic Analysis Reveals Downstream PKA Effectors of AKAP Cypher/ZASP in the Pathogenesis of Dilated Cardiomyopathy.Front Cardiovasc Med. 2021 Dec 13;8:753072. doi: 10.3389/fcvm.2021.753072. eCollection 2021. Front Cardiovasc Med. 2021. PMID: 34966794 Free PMC article.
-
The Critical Role of Oxidative Stress in Sarcopenic Obesity.Oxid Med Cell Longev. 2021 Oct 12;2021:4493817. doi: 10.1155/2021/4493817. eCollection 2021. Oxid Med Cell Longev. 2021. PMID: 34676021 Free PMC article. Review.
-
Aging, Osteo-Sarcopenia, and Musculoskeletal Mechano-Transduction.Front Rehabil Sci. 2021;2:782848. doi: 10.3389/fresc.2021.782848. Epub 2021 Dec 6. Front Rehabil Sci. 2021. PMID: 36004321 Free PMC article.
References
-
- Pisot R., Marusic U., Biolo G., Mazzucco S., Lazzer S., Grassi B., Reggiani C., Toniolo L., di Prampero P.E., Passaro A., et al. Greater loss in muscle mass and function but smaller metabolic alterations in older compared with younger men following 2 wk of bed rest and recovery. J. Appl. Physiol. 2016;120:922–929. doi: 10.1152/japplphysiol.00858.2015. - DOI - PubMed
-
- Lang F., Khaghani S., Turk C., Wiederstein J.L., Holper S., Piller T., Nogara L., Blaauw B., Gunther S., Muller S., et al. Single Muscle Fiber Proteomics Reveals Distinct Protein Changes in Slow and Fast Fibers during Muscle Atrophy. J. Proteome Res. 2018;17:3333–3347. doi: 10.1021/acs.jproteome.8b00093. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

