Widespread occurrence of microRNA-mediated target cleavage on membrane-bound polysomes
- PMID: 33402203
- PMCID: PMC7784310
- DOI: 10.1186/s13059-020-02242-6
Widespread occurrence of microRNA-mediated target cleavage on membrane-bound polysomes
Abstract
Background: Small RNAs (sRNAs) including microRNAs (miRNAs) and small interfering RNAs (siRNAs) serve as core players in gene silencing at transcriptional and post-transcriptional levels in plants, but their subcellular localization has not yet been well studied, thus limiting our mechanistic understanding of sRNA action.
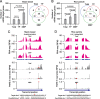
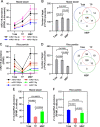
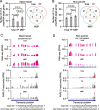
Results: We investigate the cytoplasmic partitioning of sRNAs and their targets globally in maize (Zea mays, inbred line "B73") and rice (Oryza sativa, cv. "Nipponbare") by high-throughput sequencing of polysome-associated sRNAs and 3' cleavage fragments, and find that both miRNAs and a subset of 21-nucleotide (nt)/22-nt siRNAs are enriched on membrane-bound polysomes (MBPs) relative to total polysomes (TPs) across different tissues. Most of the siRNAs are generated from transposable elements (TEs), and retrotransposons positively contributed to MBP overaccumulation of 22-nt TE-derived siRNAs (TE-siRNAs) as opposed to DNA transposons. Widespread occurrence of miRNA-mediated target cleavage is observed on MBPs, and a large proportion of these cleavage events are MBP-unique. Reproductive 21PHAS (21-nt phasiRNA-generating) and 24PHAS (24-nt phasiRNA-generating) precursors, which were commonly considered as noncoding RNAs, are bound by polysomes, and high-frequency cleavage of 21PHAS precursors by miR2118 and 24PHAS precursors by miR2275 is further detected on MBPs. Reproductive 21-nt phasiRNAs are enriched on MBPs as opposed to TPs, whereas 24-nt phasiRNAs are nearly completely devoid of polysome occupancy.
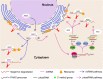
Conclusions: MBP overaccumulation is a conserved pattern for cytoplasmic partitioning of sRNAs, and endoplasmic reticulum (ER)-bound ribosomes function as an independent regulatory layer for miRNA-induced gene silencing and reproductive phasiRNA biosynthesis in maize and rice.
Keywords: Membrane-bound polysome; Oryza sativa; PARE; Target cleavage; Zea mays; miRNA; phasiRNA; siRNA.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

