Microglial gene signature reveals loss of homeostatic microglia associated with neurodegeneration of Alzheimer's disease
- PMID: 33402227
- PMCID: PMC7786928
- DOI: 10.1186/s40478-020-01099-x
Microglial gene signature reveals loss of homeostatic microglia associated with neurodegeneration of Alzheimer's disease
Abstract
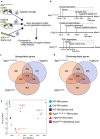
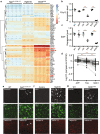
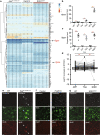
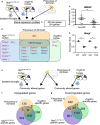
Microglia-mediated neuroinflammation has been implicated in the pathogenesis of Alzheimer's disease (AD). Although microglia in aging and neurodegenerative disease model mice show a loss of homeostatic phenotype and activation of disease-associated microglia (DAM), a correlation between those phenotypes and the degree of neuronal cell loss has not been clarified. In this study, we performed RNA sequencing of microglia isolated from three representative neurodegenerative mouse models, AppNL-G-F/NL-G-F with amyloid pathology, rTg4510 with tauopathy, and SOD1G93A with motor neuron disease by magnetic activated cell sorting. In parallel, gene expression patterns of the human precuneus with early Alzheimer's change (n = 11) and control brain (n = 14) were also analyzed by RNA sequencing. We found that a substantial reduction of homeostatic microglial genes in rTg4510 and SOD1G93A microglia, whereas DAM genes were uniformly upregulated in all mouse models. The reduction of homeostatic microglial genes was correlated with the degree of neuronal cell loss. In human precuneus with early AD pathology, reduced expression of genes related to microglia- and oligodendrocyte-specific markers was observed, although the expression of DAM genes was not upregulated. Our results implicate a loss of homeostatic microglial function in the progression of AD and other neurodegenerative diseases. Moreover, analyses of human precuneus also suggest loss of microglia and oligodendrocyte functions induced by early amyloid pathology in human.
Keywords: Alzheimer’s disease; Animal model; Microglia; Neuroinflammation; Next generation sequence; Precuneus.
Conflict of interest statement
The authors report no biomedical financial interests or potential conflicts of interest.
Figures


References
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. doi: 10.2307/2346101. - DOI
-
- Castillo E, Leon J, Mazzei G, Abolhassani N, Haruyama N, Saito T, Saido T, Hokama M, Iwaki T, Ohara T, et al. Comparative profiling of cortical gene expression in Alzheimer’s disease patients and mouse models demonstrates a link between amyloidosis and neuroinflammation. Sci Rep. 2017;7:17762. doi: 10.1038/s41598-017-17999-3. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

