Characterization of a Four-Component Regulatory System Controlling Bacteriocin Production in Streptococcus gallolyticus
- PMID: 33402539
- PMCID: PMC8545106
- DOI: 10.1128/mBio.03187-20
Characterization of a Four-Component Regulatory System Controlling Bacteriocin Production in Streptococcus gallolyticus
Abstract
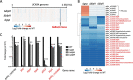
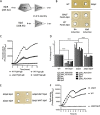
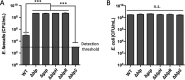
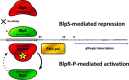
Bacteriocins are natural antimicrobial peptides produced by bacteria to kill closely related competitors. The opportunistic pathogen Streptococcus gallolyticus subsp. gallolyticus was recently shown to outcompete commensal enterococci of the murine microbiota under tumoral conditions thanks to the production of a two-peptide bacteriocin named gallocin. Here, we identified four genes involved in the regulatory control of gallocin in S. gallolyticus subsp. gallolyticus UCN34 that encode a histidine kinase/response regulator two-component system (BlpH/BlpR), a secreted peptide (GSP [gallocin-stimulating peptide]), and a putative regulator of unknown function (BlpS). While BlpR is a typical 243-amino-acid (aa) response regulator possessing a phospho-receiver domain and a LytTR DNA-binding domain, BlpS is a 108-aa protein containing only a LytTR domain. Our results showed that the secreted peptide GSP activates the dedicated two-component system BlpH/BlpR to induce gallocin transcription. A genome-wide transcriptome analysis indicates that this regulatory system (GSP-BlpH/BlpR) is specific for bacteriocin production. Importantly, as opposed to BlpR, BlpS was shown to repress gallocin gene transcription. A conserved operator DNA sequence of 30 bp was found in all promoter regions regulated by BlpR and BlpS. Electrophoretic mobility shift assays (EMSA) and footprint assays showed direct and specific binding of BlpS and BlpR to various regulated promoter regions in a dose-dependent manner on this conserved sequence. Gallocin expression appears to be tightly controlled in S. gallolyticus subsp. gallolyticus by quorum sensing and antagonistic activity of 2 LytTR-containing proteins. Competition experiments in gut microbiota medium and 5% CO2 to mimic intestinal conditions demonstrate that gallocin is functional under these in vivo-like conditions.IMPORTANCEStreptococcus gallolyticus subsp. gallolyticus, formerly known as Streptococcus bovis biotype I, is an opportunistic pathogen causing septicemia and endocarditis in the elderly often associated with asymptomatic colonic neoplasia. Recent studies indicate that S. gallolyticus subsp. gallolyticus is both a driver and a passenger of colorectal cancer. We previously showed that S. gallolyticus subsp. gallolyticus produces a bacteriocin, termed gallocin, enabling colonization of the colon under tumoral conditions by outcompeting commensal members of the murine microbiota such as Enterococcus faecalis Here, we identified and extensively characterized a four-component system that regulates gallocin production. Gallocin gene transcription is activated by a secreted peptide pheromone (GSP) and a two-component signal transduction system composed of a transmembrane histidine kinase receptor (BlpH) and a cytosolic response regulator (BlpR). Finally, a DNA-binding protein (BlpS) was found to repress gallocin genes transcription, likely by antagonizing BlpR. Understanding gallocin regulation is crucial to prevent S. gallolyticus subsp. gallolyticus colon colonization under tumoral conditions.
Keywords: Streptococcus; bacteriocins; regulation of gene expression.
Copyright © 2021 Proutière et al.
Figures




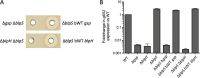
References
-
- Hoen B, Chirouze C, Cabell CH, Selton-Suty C, Duchêne F, Olaison L, Miro JM, Habib G, Abrutyn E, Eykyn S, Bernard Y, Marco F, Corey GR, International Collaboration on Endocarditis Study Group. 2005. Emergence of endocarditis due to group D streptococci: findings derived from the merged database of the International Collaboration on Endocarditis. Eur J Clin Microbiol Infect Dis 24:12–16. doi:10.1007/s10096-004-1266-6. - DOI - PubMed
-
- Corredoira-Sánchez J, García-Garrote F, Rabuñal R, López-Roses L, García-País MJ, Castro E, González-Soler R, Coira A, Pita J, López-Álvarez MJ, Alonso MP, Varela J. 2012. Association between bacteremia due to Streptococcus gallolyticus subsp. gallolyticus (Streptococcus bovis I) and colorectal neoplasia: a case-control Study. Clin Infect Dis 55:491–496. doi:10.1093/cid/cis434. - DOI - PubMed
-
- Aymeric L, Donnadieu F, Mulet C, Du Merle L, Nigro G, Saffarian A, Bérard M, Poyart C, Robine S, Regnault B, Trieu-Cuot P, Sansonetti PJ, Dramsi S. 2018. Colorectal cancer specific conditions promote Streptococcus gallolyticus gut colonization. Proc Natl Acad Sci U S A 115:E283–E291. doi:10.1073/pnas.1715112115. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
