Suppression gene drive in continuous space can result in unstable persistence of both drive and wild-type alleles
- PMID: 33404162
- PMCID: PMC7887089
- DOI: 10.1111/mec.15788
Suppression gene drive in continuous space can result in unstable persistence of both drive and wild-type alleles
Abstract
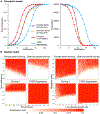
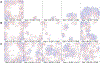
Rapid evolutionary processes can produce drastically different outcomes when studied in panmictic population models vs. spatial models. One such process is gene drive, which describes the spread of "selfish" genetic elements through a population. Engineered gene drives are being considered for the suppression of disease vectors or invasive species. While laboratory experiments and modelling in panmictic populations have shown that such drives can rapidly eliminate a population, it remains unclear if these results translate to natural environments where individuals inhabit a continuous landscape. Using spatially explicit simulations, we show that the release of a suppression drive can result in what we term "chasing" dynamics, in which wild-type individuals recolonize areas where the drive has locally eliminated the population. Despite the drive subsequently reconquering these areas, complete population suppression often fails to occur or is substantially delayed. This increases the likelihood that the drive is lost or that resistance evolves. We analyse how chasing dynamics are influenced by the type of drive, its efficiency, fitness costs, and ecological factors such as the maximal growth rate of the population and levels of dispersal and inbreeding. We find that chasing is more common for lower efficiency drives when dispersal is low and that some drive mechanisms are substantially more prone to chasing behaviour than others. Our results demonstrate that the population dynamics of suppression gene drives are determined by a complex interplay of genetic and ecological factors, highlighting the need for realistic spatial modelling to predict the outcome of drive releases in natural populations.
Keywords: biotechnology; ecological genetics; genetically modified organisms; population dynamics; population ecology; population genetics - theoretical.
© 2021 John Wiley & Sons Ltd.
Figures




References
-
- Hart SP, Usinowicz J, Levine JM. The spatial scales of species coexistence. Nat Ecol Evol, 1, 1066–1073, 2017. - PubMed
-
- Barton NH, Kelleher J, Etheridge AM. A new model for extinction and recolonization in two dimensions: quantifying phylogeography. Evolution, 64, 2701–15, 2010. - PubMed
-
- Barton NH, Depaulis F, Etheridge AM. Neutral evolution in spatially continuous populations. Theor Popul Biol, 61, 31–48, 2002. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

