Viral strategies for circumventing p53: the case of severe acute respiratory syndrome coronavirus
- PMID: 33405482
- PMCID: PMC7924916
- DOI: 10.1097/CCO.0000000000000713
Viral strategies for circumventing p53: the case of severe acute respiratory syndrome coronavirus
Abstract
Purpose of review: Virtually all viruses have evolved molecular instruments to circumvent cell mechanisms that may hamper their replication, dissemination, or persistence. Among these is p53, a key gatekeeper for cell division and survival that also regulates innate immune responses. This review summarizes the strategies used by different viruses and discusses the mechanisms deployed by SARS-CoV to target p53 activities.
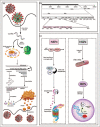
Recent findings: We propose a typology for the strategies used by different viruses to address p53 functions: hit and run (e.g. IAV, ZIKV), hide and seek (e.g. HIV1), kidnap and exploit (e.g. EBV, HSV1), dominate and suppress (e.g. HR HPV). We discuss the mechanisms by which SARS nsp3 protein targets p53 for degradation and we speculate on the significance for Covid-19 pathogenesis and risk of cancer.
Summary: p53 may operate as an intracellular antiviral defense mechanism. To circumvent it, SARS viruses adopt a kidnap and exploit strategy also shared by several viruses with transforming potential. This raises the question of whether SARS infections may make cells permissive to oncogenic DNA damage.
Copyright © 2021 Wolters Kluwer Health, Inc. All rights reserved.
Conflict of interest statement
There are no conflicts of interest.
Figures

References
-
- Wiersinga WJ, Rhodes A, Cheng AC, et al. . Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA 2020; 324:782–793. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

