Comprehensive genomic analysis reveals virulence factors and antibiotic resistance genes in Pantoea agglomerans KM1, a potential opportunistic pathogen
- PMID: 33406073
- PMCID: PMC7787473
- DOI: 10.1371/journal.pone.0239792
Comprehensive genomic analysis reveals virulence factors and antibiotic resistance genes in Pantoea agglomerans KM1, a potential opportunistic pathogen
Abstract
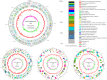
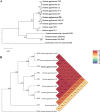
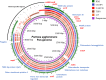
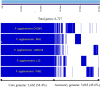
Pantoea agglomerans is a Gram-negative facultative anaerobic bacillus causing a wide range of opportunistic infections in humans including septicemia, pneumonia, septic arthritis, wound infections and meningitis. To date, the determinants of virulence, antibiotic resistance, metabolic features conferring survival and host-associated pathogenic potential of this bacterium remain largely underexplored. In this study, we sequenced and assembled the whole-genome of P. agglomerans KM1 isolated from kimchi in South Korea. The genome contained one circular chromosome of 4,039,945 bp, 3 mega plasmids, and 2 prophages. The phage-derived genes encoded integrase, lysozyme and terminase. Six CRISPR loci were identified within the bacterial chromosome. Further in-depth analysis showed that the genome contained 13 antibiotic resistance genes conferring resistance to clinically important antibiotics such as penicillin G, bacitracin, rifampicin, vancomycin, and fosfomycin. Genes involved in adaptations to environmental stress were also identified which included factors providing resistance to osmotic lysis, oxidative stress, as well as heat and cold shock. The genomic analysis of virulence factors led to identification of a type VI secretion system, hemolysin, filamentous hemagglutinin, and genes involved in iron uptake and sequestration. Finally, the data provided here show that, the KM1 isolate exerted strong immunostimulatory properties on RAW 264.7 macrophages in vitro. Stimulated cells produced Nitric Oxide (NO) and pro-inflammatory cytokines TNF-α, IL-6 and the anti-inflammatory cytokine IL-10. The upstream signaling for production of TNF-α, IL-6, IL-10, and NO depended on TLR4 and TLR1/2. While production of TNF-α, IL-6 and NO involved solely activation of the NF-κB, IL-10 secretion was largely dependent on NF-κB and to a lesser extent on MAPK Kinases. Taken together, the analysis of the whole-genome and immunostimulatory properties provided in-depth characterization of the P. agglomerans KM1 isolate shedding a new light on determinants of virulence that drive its interactions with the environment, other microorganisms and eukaryotic hosts.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Comparative genomics to examine the endophytic potential of Pantoea agglomerans DAPP-PG 734.BMC Genomics. 2022 Nov 8;23(1):742. doi: 10.1186/s12864-022-08966-y. BMC Genomics. 2022. PMID: 36344949 Free PMC article.
-
Polyphasic study of plant- and clinic-associated Pantoea agglomerans strains reveals indistinguishable virulence potential.Infect Genet Evol. 2009 Dec;9(6):1381-91. doi: 10.1016/j.meegid.2009.09.016. Epub 2009 Oct 2. Infect Genet Evol. 2009. PMID: 19800991
-
Pantoea agglomerans: a mysterious bacterium of evil and good. Part III. Deleterious effects: infections of humans, animals and plants.Ann Agric Environ Med. 2016 Jun 2;23(2):197-205. doi: 10.5604/12321966.1203878. Ann Agric Environ Med. 2016. PMID: 27294620 Review.
-
Phylogeny and identification of Pantoea species and typing of Pantoea agglomerans strains by multilocus gene sequencing.J Clin Microbiol. 2009 Feb;47(2):300-10. doi: 10.1128/JCM.01916-08. Epub 2008 Dec 3. J Clin Microbiol. 2009. PMID: 19052179 Free PMC article.
-
Virulence mechanisms and host specificity of gall-forming Pantoea agglomerans.Trends Microbiol. 2007 Dec;15(12):538-45. doi: 10.1016/j.tim.2007.10.009. Epub 2007 Nov 19. Trends Microbiol. 2007. PMID: 18024130 Review.
Cited by
-
Safety of grain and flour from perennial intermediate wheatgrass (Thinopyrum intermedium) as a novel food pursuant to Regulation (EU) 2015/2283.EFSA J. 2025 Jun 10;23(6):e9467. doi: 10.2903/j.efsa.2025.9467. eCollection 2025 Jun. EFSA J. 2025. PMID: 40496252 Free PMC article.
-
An Emerging Bacterial Leaf Disease in Rice Caused by Pantoea ananatis and Pantoea eucalypti in Northeast China.Microorganisms. 2025 Jun 13;13(6):1376. doi: 10.3390/microorganisms13061376. Microorganisms. 2025. PMID: 40572264 Free PMC article.
-
Unveiling the prevalence and antimicrobial resistance landscape of Pantoea genus in veterinary clinical strains: Insights from a cohort study.One Health. 2025 Jul 7;21:101130. doi: 10.1016/j.onehlt.2025.101130. eCollection 2025 Dec. One Health. 2025. PMID: 40704225 Free PMC article.
-
Comparative genomics to examine the endophytic potential of Pantoea agglomerans DAPP-PG 734.BMC Genomics. 2022 Nov 8;23(1):742. doi: 10.1186/s12864-022-08966-y. BMC Genomics. 2022. PMID: 36344949 Free PMC article.
-
Opposite Sides of Pantoea agglomerans and Its Associated Commercial Outlook.Microorganisms. 2022 Oct 20;10(10):2072. doi: 10.3390/microorganisms10102072. Microorganisms. 2022. PMID: 36296348 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

