Probabilistic models of biological enzymatic polymerization
- PMID: 33406128
- PMCID: PMC7787436
- DOI: 10.1371/journal.pone.0244858
Probabilistic models of biological enzymatic polymerization
Abstract
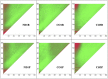
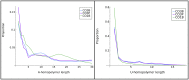
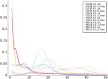
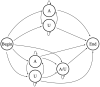
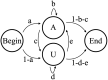
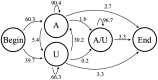
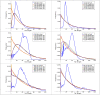
In this study, hierarchies of probabilistic models are evaluated for their ability to characterize the untemplated addition of adenine and uracil to the 3' ends of mitochondrial mRNAs of the human pathogen Trypanosoma brucei, and for their generative abilities to reproduce populations of these untemplated adenine/uridine "tails". We determined the most ideal Hidden Markov Models (HMMs) for this biological system. While our HMMs were not able to generatively reproduce the length distribution of the tails, they fared better in reproducing nucleotide composition aspects of the tail populations. The HMMs robustly identified distinct states of nucleotide addition that correlate to experimentally verified tail nucleotide composition differences. However they also identified a surprising subclass of tails among the ND1 gene transcript populations that is unexpected given the current idea of sequential enzymatic action of untemplated tail addition in this system. Therefore, these models can not only be utilized to reflect biological states that we already know about, they can also identify hypotheses to be experimentally tested. Finally, our HMMs supplied a way to correct a portion of the sequencing errors present in our data. Importantly, these models constitute rare simple pedagogical examples of applied bioinformatic HMMs, due to their binary emissions.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

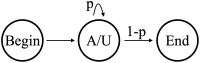

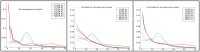
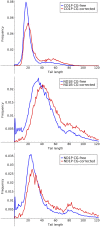


References
-
- Durbin R, Eddy SR, Krogh A, Mitchison GJ. Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids. Cambridge University Press, Cambridge UK; 1998.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

