Genetic Variation and Evolution of the 2019 Novel Coronavirus
- PMID: 33406522
- PMCID: PMC7900485
- DOI: 10.1159/000513530
Genetic Variation and Evolution of the 2019 Novel Coronavirus
Abstract
Introduction: SARS-CoV-2 is a new type of coronavirus causing a pandemic severe acute respiratory syndrome (SARS-2). Coronaviruses are very diverting genetically and mutate so often periodically. The natural selection of viral mutations may cause host infection selectivity and infectivity.
Methods: This study was aimed to indicate the diversity between human and animal coronaviruses through finding the rate of mutation in each of the spike, nucleocapsid, envelope, and membrane proteins.
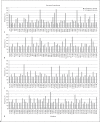
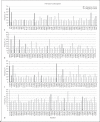
Results: The mutation rate is abundant in all 4 structural proteins. The most number of statistically significant amino acid mutations were found in spike receptor-binding domain (RBD) which may be because it is responsible for a corresponding receptor binding in a broad range of hosts and host selectivity to infect. Among 17 previously known amino acids which are important for binding of spike to angiotensin-converting enzyme 2 (ACE2) receptor, all of them are conservative among human coronaviruses, but only 3 of them significantly are mutated in animal coronaviruses. A single amino acid aspartate-454, that causes dissociation of the RBD of the spike and ACE2, and F486 which gives the strength of binding with ACE2 remain intact in all coronaviruses.
Discussion/conclusion: Observations of this study provided evidence of the genetic diversity and rapid evolution of SARS-CoV-2 as well as other human and animal coronaviruses.
Keywords: Biodiversity; Clinical genetics; Ecology; Mutation rate; Pandemics; Prevalence.
© 2021 S. Karger AG, Basel.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

