Ever-increasing viral diversity associated with the red imported fire ant Solenopsis invicta (Formicidae: Hymenoptera)
- PMID: 33407622
- PMCID: PMC7788728
- DOI: 10.1186/s12985-020-01469-w
Ever-increasing viral diversity associated with the red imported fire ant Solenopsis invicta (Formicidae: Hymenoptera)
Abstract
Background: Advances in sequencing and analysis tools have facilitated discovery of many new viruses from invertebrates, including ants. Solenopsis invicta is an invasive ant that has quickly spread worldwide causing significant ecological and economic impacts. Its virome has begun to be characterized pertaining to potential use of viruses as natural enemies. Although the S. invicta virome is the best characterized among ants, most studies have been performed in its native range, with less information from invaded areas.
Methods: Using a metatranscriptome approach, we further identified and molecularly characterized virus sequences associated with S. invicta, in two introduced areas, U.S and Taiwan. The data set used here was obtained from different stages (larvae, pupa, and adults) of S. invicta life cycle. Publicly available RNA sequences from GenBank's Sequence Read Archive were downloaded and de novo assembled using CLC Genomics Workbench 20.0.1. Contigs were compared against the non-redundant protein sequences and those showing similarity to viral sequences were further analyzed.
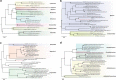
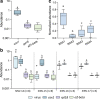
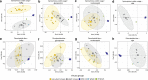
Results: We characterized five putative new viruses associated with S. invicta transcriptomes. Sequence comparisons revealed extensive divergence across ORFs and genomic regions with most of them sharing less than 40% amino acid identity with those closest homologous sequences previously characterized. The first negative-sense single-stranded RNA virus genomic sequences included in the orders Bunyavirales and Mononegavirales are reported. In addition, two positive single-strand virus genome sequences and one single strand DNA virus genome sequence were also identified. While the presence of a putative tenuivirus associated with S. invicta was previously suggested to be a contamination, here we characterized and present strong evidence that Solenopsis invicta virus 14 (SINV-14) is a tenui-like virus that has a long-term association with the ant. Furthermore, based on virus sequence abundance compared to housekeeping genes, phylogenetic relationships, and completeness of viral coding sequences, our results suggest that four of five virus sequences reported, those being SINV-14, SINV-15, SINV-16 and SINV-17, may be associated to viruses actively replicating in the ant S. invicta.
Conclusions: The present study expands our knowledge about viral diversity associated with S. invicta in introduced areas with potential to be used as biological control agents, which will require further biological characterization.
Keywords: Bunyavirales; Metatranscriptome; Mononegavirales; Red imported fire ant; S. invicta; Tenuivirus; Virome; Virus diversity.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

