Phosphorylation of Influenza A Virus NS1 at Serine 205 Mediates Its Viral Polymerase-Enhancing Function
- PMID: 33408177
- PMCID: PMC8094967
- DOI: 10.1128/JVI.02369-20
Phosphorylation of Influenza A Virus NS1 at Serine 205 Mediates Its Viral Polymerase-Enhancing Function
Abstract
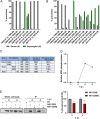
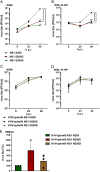
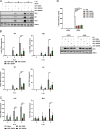
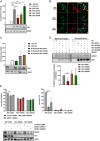
Influenza A virus (IAV) nonstructural protein 1 (NS1) is a protein with multiple functions that are regulated by phosphorylation. Phosphoproteomic screening of H1N1 virus-infected cells revealed that NS1 was phosphorylated at serine 205 in intermediate stages of the viral life cycle. Interestingly, S205 is one of six amino acid changes in NS1 of post-pandemic H1N1 viruses currently circulating in humans compared to the original swine-origin 2009 pandemic (H1N1pdm09) virus, suggesting a role in host adaptation. To identify NS1 functions regulated by S205 phosphorylation, we generated recombinant PR8 H1N1 NS1 mutants with S205G (nonphosphorylatable) or S205N (H1N1pdm09 signature), as well as H1N1pdm09 viruses harboring the reverse mutation NS1 N205S or N205D (phosphomimetic). Replication of PR8 NS1 mutants was attenuated relative to wild-type (WT) virus replication in a porcine cell line. However, PR8 NS1 S205N showed remarkably higher attenuation than PR8 NS1 S205G in a human cell line, highlighting a potential host-independent advantage of phosphorylatable S205, while an asparagine at this position led to a potential host-specific attenuation. Interestingly, PR8 NS1 S205G did not show polymerase activity-enhancing functions, in contrast to the WT, which can be attributed to diminished interaction with cellular restriction factor DDX21. Analysis of the respective kinase mediating S205 phosphorylation indicated an involvement of casein kinase 2 (CK2). CK2 inhibition significantly reduced the replication of WT viruses and decreased NS1-DDX21 interaction, as observed for NS1 S205G. In summary, NS1 S205 is required for efficient NS1-DDX21 binding, resulting in enhanced viral polymerase activity, which is likely to be regulated by transient phosphorylation.IMPORTANCE Influenza A viruses (IAVs) still pose a major threat to human health worldwide. As a zoonotic virus, IAV can spontaneously overcome species barriers and even reside in new hosts after efficient adaptation. Investigation of the functions of specific adaptational mutations can lead to a deeper understanding of viral replication in specific hosts and can probably help to find new targets for antiviral intervention. In the present study, we analyzed the role of NS1 S205, a phosphorylation site that was reacquired during the circulation of pandemic H1N1pdm09 "swine flu" in the human host. We found that phosphorylation of human H1N1 virus NS1 S205 is mediated by the cellular kinase CK2 and is needed for efficient interaction with human host restriction factor DDX21, mediating NS1-induced enhancement of viral polymerase activity. Therefore, targeting CK2 activity might be an efficient strategy for limiting the replication of IAVs circulating in the human population.
Keywords: NS1; influenza virus; protein phosphorylation; viral polymerase activity.
Copyright © 2021 American Society for Microbiology.
Figures
References
-
- Mok BW, Song W, Wang P, Tai H, Chen Y, Zheng M, Wen X, Lau SY, Wu WL, Matsumoto K, Yuen KY, Chen H. 2012. The NS1 protein of influenza A virus interacts with cellular processing bodies and stress granules through RNA-associated protein 55 (RAP55) during virus infection. J Virol 86:12695–12707. doi:10.1128/JVI.00647-12. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

