Large-scale, three-dimensional tissue cytometry of the human kidney: a complete and accessible pipeline
- PMID: 33408350
- PMCID: PMC8363780
- DOI: 10.1038/s41374-020-00518-w
Large-scale, three-dimensional tissue cytometry of the human kidney: a complete and accessible pipeline
Abstract
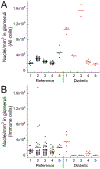
The advent of personalized medicine has driven the development of novel approaches for obtaining detailed cellular and molecular information from clinical tissue samples. Tissue cytometry is a promising new technique that can be used to enumerate and characterize each cell in a tissue and, unlike flow cytometry and other single-cell techniques, does so in the context of the intact tissue, preserving spatial information that is frequently crucial to understanding a cell's physiology, function, and behavior. However, the wide-scale adoption of tissue cytometry as a research tool has been limited by the fact that published examples utilize specialized techniques that are beyond the capabilities of most laboratories. Here we describe a complete and accessible pipeline, including methods of sample preparation, microscopy, image analysis, and data analysis for large-scale three-dimensional tissue cytometry of human kidney tissues. In this workflow, multiphoton microscopy of unlabeled tissue is first conducted to collect autofluorescence and second-harmonic images. The tissue is then labeled with eight fluorescent probes, and imaged using spectral confocal microscopy. The raw 16-channel images are spectrally deconvolved into 8-channel images, and analyzed using the Volumetric Tissue Exploration and Analysis (VTEA) software developed by our group. We applied this workflow to analyze millimeter-scale tissue samples obtained from human nephrectomies and from renal biopsies from individuals diagnosed with diabetic nephropathy, generating a quantitative census of tens of thousands of cells in each. Such analyses can provide useful insights that can be linked to the biology or pathology of kidney disease. The approach utilizes common laboratory techniques, is compatible with most commercially-available confocal microscope systems and all image and data analysis is conducted using the VTEA image analysis software, which is available as a plug-in for ImageJ.
Conflict of interest statement
Disclosures/Conflicts of Interest
The authors declare that they have no conflicts of interest
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

