Determinants of dengue virus dispersal in the Americas
- PMID: 33408877
- PMCID: PMC7772473
- DOI: 10.1093/ve/veaa074
Determinants of dengue virus dispersal in the Americas
Abstract
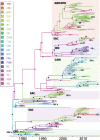
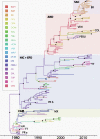
Dengue viruses (DENVs) are classified into four serotypes, each of which contains multiple genotypes. DENV genotypes introduced into the Americas over the past five decades have exhibited different rates and patterns of spatial dispersal. In order to understand factors underlying these patterns, we utilized a statistical framework that allows for the integration of ecological, socioeconomic, and air transport mobility data as predictors of viral diffusion while inferring the phylogeographic history. Predictors describing spatial diffusion based on several covariates were compared using a generalized linear model approach, where the support for each scenario and its contribution is estimated simultaneously from the data set. Although different predictors were identified for different serotypes, our analysis suggests that overall diffusion of DENV-1, -2, and -3 in the Americas was associated with airline traffic. The other significant predictors included human population size, the geographical distance between countries and between urban centers and the density of people living in urban environments.
Keywords: Bayesian phylogeography; dengue virus; hypothesis testing; spatial diffusion.
© The Author(s) 2020. Published by Oxford University Press.
Figures




References
-
- Bennett S. N. (2006) ‘Molecular Evolution of Dengue 2 Virus in Puerto Rico: positive Selection in the Viral Envelope Accompanies Clade Reintroduction’, Journal of General Virology, 87: 885–93. - PubMed

